Purpose:
- To see DNA by lysing (breaking open) bacterial cells in an alkaline solution and releasing a viscous thread of DNA that can be lifted from the slide surface with a toothpick.
- To determine the Gram-stain reaction of bacterial isolates by attempting to lyse the bacteria in 3% (w/v) potassium hydroxide (KOH). Only Gram-negative cells will lyse.
Background Information:
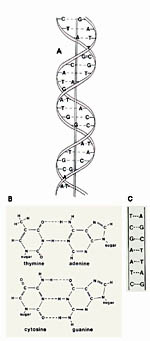
All students need to be familiar with basic cell structure including cell walls and the presence of deoxyribonucleic acid (DNA) in the cell. DNA is a very long, double-stranded, helical molecule composed of many nucleotide units. Each unit is composed of a sugar (deoxyribose), a phosphate group, and a nitrogenous base. Four nitrogenous bases are found in DNA: the purines, adenine (A) and guanine (G), and the pyrimidines, cytosine (C) and thymine (T). (FIGURE 1) In the double strand of DNA, the nitrogenous bases pair in a distinct pattern; A always pairs with T, and G always pairs with C, to form the rungs of the DNA helical ladder. This nucleotide pairing aids in the rapid and accurate replication of the DNA double helix; each time cells divide, the DNA is copied, or replicated.
|
|
Figure 1.
The double helix. a. Deoxyribonucleic acid (DNA) is a double-stranded, helical molecule composed of nucleotide units. Replication of DNA occurs through the pairing of nucleotide bases to form new strands. b. Diagram of the molecular structure of the base pairs of DNA. “Sugar” indicates the bond to deoxyribose. Phosphates connect the sugars to make the “backbone” of DNA. A nitrogen base plus deoxyribose and a phosphate group constitutes a nucleotide. Adenine (A) always pairs with thymine (T), and cytosine (C) always pairs with guanine (G). c. Diagram showing pairing of the nucleotide bases in a short DNA segment. (Courtesy G.L. Schumann). Click image to see a larger view. |
The genetic code is interpreted through the production of proteins, many of which are enzymes that control cell functions. A gene is a segment of DNA that encodes a product, usually a protein. DNA is transcribed into messenger RNA (ribonuleic acid), a closely related molecule. Transfer RNAs carry amino acids to the messenger RNA where they are lined up in the correct order to build a protein which completes the translation of the original DNA genetic code. This process occurs within ribosomes, composed partially of the third type of RNA, ribosomal RNA. (FIGURE 2)
 |
Figure 2.
The translation of a deoxyribonucleic acid (DNA) genetic message into a protein. Ribonucleic acid (RNA) differs from DNA in having ribose sugar units rather than deoxyribose and uracil (U) rather than thymine (T) as a nucleotide base. a. Overview of transcription using one strand of DNA to messenger RNA (mRNA) and translation to a chain of amino acids (protein) with the help of transfer RNAs (tRNAs). b. After transcription from DNA, mRNA leaves the nucleus and moves to the cytoplasm in eukaryotes. tRNAs carry amino acids and “match” with a triplet codon on the mRNA to create a chain of amino acids in the order coded for in the DNA.
c. Diagrammatic view of transcription and translation. Nucleotide bases: C=cytosine, G=guanine, A=adenine. Amino acids: ser=serine, tyr=tyrosine, arg=arginine. (Courtesy G. Schumann). Click image to see a larger view. |
It is beyond the scope of this exercise to describe the details of DNA function and the genetic code, but this information is found in all general biology books. Diagrams are included in this exercise for classroom use.
DNA is the basis of life because it contains the genetic code for all living organisms. The genes contained in an organism’s DNA make up the blueprint for that organism, and determine its appearance and all of its functions. Chromosomes are composed primarily of long, thin strands of DNA. In eukaryotic organisms, such as plants, animals and fungi, the chromosomes are contained in a membrane-bound nucleus. In prokaryotes, such as bacteria, the single chromosome is in the cytoplasm and may be accompanied by smaller pieces of circular DNA called plasmids.
Biotechnology and genetic engineering also are common topics in many classrooms. The first step in molecular biology and genetic engineering is to isolate DNA from cells. Because the genetic code of DNA is nearly universal, any gene can potentially be transferred from a donor organism and function in a related or unrelated recipient organism. The rapid and simple visualization of the stringing effect of DNA from lysed bacterial cells in this exercise may help attract the interest of students to these subjects. This technique is similar to several others that are commonly used to isolate DNA from cells of plants, such as onions.
This diagram illustrates a bacterial cell with a single large chromosome and an extra circular piece of DNA known as a plasmid (FIGURE 3).
 |
Figure 3.
Agrobacterium tumefaciens, the crown gall bacterium, a natural genetic engineer. (Used by permission of P. Sforza). Click image to see a larger view. |
The bacterial chromosome and bacterial plasmids were visualized in an electron microscope, after they were released from an unidentified bacterial cell (FIGURE 4). The cell was broken gently so that the DNA would remain intact. The plasmids (see arrows) are the circular structures that are much smaller than the main chromosomal DNA.
 |
Figure 4.
Release of chromosomal and plasmid (arrows) DNA from an unidentified bacterium. (Courtesy H. Potter and D. Dressler, from Brock Biology of Microorganisms, 9th edition, used by permission of M. T. Madigan). Click image to see a larger vie. |
To prepare students for this demonstration, a teacher has suggested that students could use plastic eggs (to represent the bacterial cells) containing string (to represent the DNA). A chopstick (toothpick) could be used to pick up the string (DNA) only when the egg is broken open, and the string is released.
In more advanced classes, the study of the biology of bacteria requires some knowledge of the Gram-stain reaction. This technique is essential in the identification and classification of bacteria. In 1884, Hans Christian Gram found that bacteria could be divided into two groups- those that retained the crystal violet stain solution with which he was experimenting (Gram-positive bacteria) and those that did not (Gram-negative). Although Gram did not appreciate the significance of this reaction at the time, we now know that this differential reaction reflects a difference in the cell wall structure and separates bacteria into two groups that reflect their evolutionary separation long ago.
Gram-positive bacteria have a cell wall composed of a single thick layer of peptidoglycan. Gram-negative bacteria have a cell wall with several thinner layers composed of peptidoglycan, lipopolysaccharide, and protein. (FIGURE 5)
 |
Figure 5.
Diagrams of the cell wall structure of Gram-negative (left) and Gram-positive bacteria. (Used by permission of P. Sforza). Click image to see a larger view. |
The Gram-stain reaction can identify bacteria as Gram-positive or Gram-negative, but potentially messy stains and expensive microscopes are needed. The KOH test is a faster and simpler method to determine the same reaction. When Gram-negative bacterial cells are placed in an alkaline solution (3% KOH), the cells walls are destroyed, and the cell contents, including the DNA, are released. Because their cell wall structure is different, Gram-positive bacteria will not lyse in 3% KOH. The KOH method was originally developed by a Japanese scientist named Ryu in 1938.
The Gram stain is an important diagnostic tool in human medicine because some antibiotics are effective against only Gram-negative bacteria (e.g. erythromycin) and some against only Gram-positive ones (e.g. penicillin, actinomycin). Bacteria also cause plant diseases. Most plant pathogenic bacteria are Gram-negative (Agrobacterium, Erwinia, Pseudomonas, Ralstonia, and Xanthomonas). A few bacterial genera found in association with plants are Gram-positive (Bacillus, Clavibacter, and Streptomyces).
