NOTE: This discussion is designed to provide a very broad outline of each of these two techniques. Further information should be sought from the web-links provided at the end of each section. These links also have great animations that will help with visualization of the procedures used.
Polymerase chain reaction is a method that can be used to amplify a segment of DNA without the need to use bacteria or other organisms as intermediaries (i.e. no transformation procedures are needed). It was initially developed in 1983 by Kary B. Mullis and perfected with the help of many of his colleagues. It is commonly used in a plant pathology laboratory to study genes and other areas of the plant's or pathogenic organism's DNA to determine structure and function of that region and to understand the gene's importance to the plant or pathogen. In this and other sciences, it is used for DNA fingerprinting, detection of hereditary genes, cloning of genes, paternity testing and detection of introduced or foreign DNA.
DNA polymerase is found in all living cells where it acts to replicate the DNA molecule each time the cell divides. The polymerase has the ability to work both inside of the cell and also outside, in a test tube. However, for the polymerase to help catalyze the formation of a new DNA molecule, the double stranded DNA must first be denatured. This denaturation or unwinding and separation of the two strands of the DNA helix occurs in vitro when the molecule is heated to a temperature of 90°C or more. Unfortunately, this temperature also causes normal DNA polymerase to denature and it is no longer able to function. This problem was solved with the discovery of the bacteria, Thermus aquaticus (Taq), that survive in very hot environments, specifically the hot mineral springs found in Yellowstone. The DNA polymerase of this bacteria continues to function even when it is extremely hot (more than 110°C).
Using the Taq polymerase scientists now had a method with which they could place the DNA that they wished to study into a tube along with the necessary components to manufacture new DNA. This includes Taq polymerase, the four nucleotide bases (A, C, T, G), and primers. Primers are very short pieces of DNA that are complimentary to the 3' end of each of the two DNA strands from the region of interest. These primers are small enough that they are able to rapidly find and bind to their complimentary DNA once the large DNA molecule has been denatured (the two strands of the helix separate into single strands). The polymerase attaches to these small areas of double stranded DNA where the primer has bound and begins manufacturing of the remainder of the segment strand. Generally there will be about a million copies made within 3 hours of running a PCR reaction.
|

|
| Figure 1 - Schematic drawing of the PCR cycle (click to enlarge) |
Special machines have been developed, called thermocyclers, that have an automated system that rapidly heats the DNA to denature it, cools down instantly allowing the primers to anneal to the appropriate strand, and then maintains a constant temperature (around 50-60°C) while the polymerase adds complementary bases from the end of the primer to make an exact copy of the original DNA strand. This process is then repeated many times with an exponential increase in the number of copies of DNA that are made.
 |
 |
|
Figures 2 & 3 - Examples of two thermocyclers:
Applied Biosystems 9700 GeneAmp (left) and iCycler thermocycler from BioRad (right) |
Once the process has been completed the researcher has enough copies of the DNA segment of interest that it can now be studied extensively to resolve the questions being investigated. Some of these will be related to the function of the DNA when it is in the cell and its role in other cell functions such as metabolism, cell maintenance and response to invasion by pathogens or abiotic stresses. We can also use PCR to check for the presence of an introduced gene. The primers only anneal to DNA that matches exactly. If primers are used that match the introduced DNA segment from a known donor organism, we will have a PCR-amplified product of specific size.
Description of the polymerase chain reaction (PCR) and how it is used as well as animations of the process can be found at:
http://www.accessexcellence.org/AB/IE/PCR_Xeroxing_DNA.html
http://depts.washington.edu/~genetics/courses/genet371b-aut99/PCR_contents.html
http://www.dnalc.org/ddnalc/resources/pcr.html
Microarray Technology
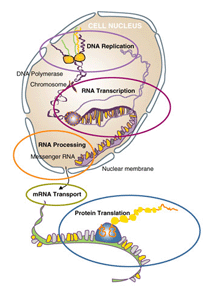
Living cells store the information to guide their functions in the form of DNA. The genetic information stored in DNA is used to direct the synthesis of the thousands of proteins each cell requires. Gene expression refers to the subset of genes within an organism that are "turned on" or active at any particular time in the cell's lifecycle. When genes are expressed, the information contained in the DNA is transcribed into RNA, which is then further processed to produce messenger RNA (mRNA). It is this mRNA molecule that moves into the cell's cytoplasm and is then translated into proteins. This translation occurs on the ribosomes present in the cytoplasm. Proteins are used by the cell for most of its critical functions.
|
|
| Figure 4 - Replication, transcription, and translation (click to enlarge) |
As the cell responds to changing environments or metabolic requirements, different proteins are needed and hence different mRNA is present within the cell. By studying the mRNAs that are present in the cell at a particular time, scientists are able to determine which genes are turned on and which are turned off at critical times in the cell's development or in response to stress. They are also able to determine the changing level of expression of genes as changes in the cell or it's environment occur.
Microarrays are designed to analyze thousands of mRNA samples at the same time. The process utilizes glass slides, silicon chips, or nylon membranes that are covered with a regular array of spots of genes from the original DNA template of the organism. It is this template from which the mRNAs were made when the cell was expressing a particular gene. Each mRNA present in the experimental sample will hybridize to the single-stranded DNA segment containing the sequence complementary to the mRNA. A fluorescent tag may be attached by amplifying the mRNA or by synthesizing cDNA from the mRNA expressed in the cell, which can then be detected and quantified by a computer. The amount of fluorescence is detected by special scanners with lasers. The signal is the amount of fluorescent light emitted. The more mRNA/cDNA present at each hybridization spot, the stronger the signal it will give. These spots are analyzed by a computer, since the difference in spot intensity is difficult to quantify in any other way. The computer measures the intensity of each spot precisely, and generates a profile of gene expression in the cell under the given experimental or natural conditions.
The data generated from a microarray experiment can tell scientists how the environment is affecting the plant. For example, different genes may be expressed when a cell is under stress from pathogen invasion or environmental changes and this change can be quantified using microarray analysis.
A discussion of microarray technology with further details and animations may be found at the following websites:
http://ceprap.ucdavis.edu/acrobat/dna_chips.pdf
http://www.ncbi.nlm.nih.gov/About/primer/microarrays.html
http://www.scq.ubc.ca/?p=272
http://www.affymetrix.com/about_affymetrix/outreach/educator/index.affx
Some of the Uses of Microarrays for Plant Pathology Include:
- understanding the effects of disease on a plant - how the plant responds.
- understanding the effects of an engineered gene on a plant.
The following web site includes an animated show that describes how DNA microarrays are produced and how they may be hybridized to answer questions about the DNA in which we are interested.
http://www.bio.davidson.edu/courses/genomics/chip/chip.html
Three other classroom activities for high school students can be found at:
http://www.scienceteacherprogram.org/biology/Meyer06.html
Student Activity: Polymerase Chain Reaction (PCR) and Microarray
- What do the initials PCR stand for?
- polymerization chain reactor
- polymerase chain reaction
- polymerization containing reaction
- polymerase chain reading
- Who invented PCR?
- Francis Crick
- Rosalind Franklin
- Albert Einstein
- Kary B. Mullis
- What is the purpose for which PCR is used?
- to amplify DNA
- to isolate DNA
- to study the function of DNA
- to observe the expression of DNA
- Which of the following best describes the mixture placed in a PCR vial at the beginning of the PCR procedure?
- the original DNA sample, ribonuclease, primers and buffer solution
- the original DNA sample, PCR enzyme, the four nucleotide bases and primers
- the original DNA sample, DNA polymerase, the four nucleotide bases and primers
- the original DNA sample, DNA polymerase, buffer
- What happens to the DNA when we heat it to 90-95°C for 30 seconds in the first step of the PCR procedure?
- the DNA strands separate
- the primers bind to the single DNA strands
- DNA polymerase extends the new strand by adding bases to the primer segments that are already attached
- the PCR process repeats itself
- What is the optimum temperature for the enzyme DNA polymerase used in the PCR process?
- 40°C
- 55°C
- 72°C
- 90°C
- How many copies of a single original DNA strand would you expect to find after about 30 cycles of the PCR process?
- 100,000
- 1,000,000
- 10,000,000
- 1,000,000,000
- A microarray is?
- a collection of microbes
- an array of microscopic holes
- a regular array of genic DNA attached to a glass, silicon or nylon chip
- a type of microscope
- What is "gene expression"?
- DNA that is known to contain genes that can be expressed
- the sequence of DNA that is related to a gene
- the production of mRNA and proteins that occurs when a gene is transcribed at certain times in the life of the cell
- the movement of genes from one cell to the other
- How do we see the hybridized spots on an array?
- the spots are just big enough for us to see
- the cDNA or mRNA probe has been labeled with a fluorescent tag that the computer can detect
- the spots turn orange
- only the hybridized spots stay on the chip
- Can we determine how much mRNA is expressed in the cell?
- Yes, because the computer is able to analyze each hybridized spot and calculate how much is present from the amount of fluorescence it detects
- No, there is way too much mRNA for us to see any differences
- No, mRNA amounts cannot be quantified
- Yes, because we can see how much brighter the spots are than ones that are not hybridized
Adapted with permission from a website by the Association of the British Pharmaceutical Industry
http://www.abpischools.org.uk/res/coResourceImport/resources/
poster-series/pcr/quiz.cfm
