Lists of New, Emerging, Re-emerging, and Threatening Plant Diseases have been developed and published in Phytopathology News (1995, Volume 29(12):217) and included in posters at meetings on several occasions. The original development of such lists came from responses to a survey sent out under the auspices of former APS President Cleo D’Arcy in 1994 for the APS National Plant Pathology Board, headed by Dr. Anne Vidaver. Lists have been prepared, corrected, updated, and placed on the Internet for all to use under the leadership of Dr. O. W. Barnett, Department of Plant Pathology, North Carolina State University (http://www.ces.ncsu.edu/index.php?page=home).
Pathogens can be classified in four categories: (1) New - pathogens detected within the last five years; (2) Emerging - pathogen incidence has increased within the last 20 years; (3) Re-emerging - pathogens associated with chemical resistance or changes in management or cultivars, previously controlled infections; and, (4) Threatening - pathogens not reported or limited in distribution in the U.S. A fifth category of ‘Chronic/spreading’ for pathogens known for longer than 20 years and causing increased concern also is included in the lists.
Our goal in preparing for this Colloquium on New and Emerging Plant Viruses has not been to formulate an all-inclusive list of potential new and emerging diseases (Table 1) but to discuss some of the underlying causes for the discovery, development, and understanding of how and why new virus diseases arise, increase in importance, invade new territories, and then wane. We trust this Colloquium will inform, generate interest, answer some questions, and create an awareness of current plant virus problems.
Plant viruses have been recognized as plant pathogens for just over 100 years and the number of recognized plant viruses has grown exponentially, in part by the discovery of new virus diseases, in part by the recognition of existing problems as virally caused diseases, in part by the origin of new virus entities and, in part by refining our techniques for separating and identifying the correct causal agents. Viruses require avenues of entry into their plant hosts and arthropods of many types play the role of vectors.
Nature requires variability for selection to occur. Variability may be driven by the host component as new resistance genes are developed against a population of vectors and viruses, by the vector component as they mutate and adapt to new host systems, or by the virus as mutations and recombinations occur. Man plays an instrumental role in genetically modifying the host plants, by eliminating weeds and insects with selective pesticides and cultural practices, by rapid dissemination of germplasm to new areas, and by inadvertently or intentionally introducing vectors, viruses, and potential hosts into existing crop systems. These combinations may be detrimental to survival of the viral component or expand opportunities for increase. Examples include: 1) cacao swollen shoot in Africa after cacao was introduced into an environment where the virus had existed in equilibrium with native species; and 2) soybean dwarf in Northern Japan after soybeans replaced rice over large areas of Hokkaido bringing together a susceptible host with an aphid biotype which preferred it and a virus which had been in equilibrium with native legumes.
Not all "new" virus diseases are caused by new viruses or new virus strains but are regarded as new diseases when they appear after a vector species or biotype or a virus is introduced into a new area. One of the virus groups which has literally exploded onto the scene in the last ten years are the Geminiviruses, specifically that group of geminiviruses transmitted by whiteflies. The introduction and spread of a new biotype of
into new areas has been directly correlated with the increase in new and different geminivirus diseases in tomato, cotton, melons, lettuce, beans, and other crops.
group are restricted to the tropics, sub-tropics, or warm temperate climes as are the diseases they transmit in nature. The next group of viruses, the thrip-vectored tosposviruses, are not restricted by temperature and have become major problems in recent years. This group has a long history as causal agents of diseases of a wide range of ornamentals, vegetables, and floral crops. Tospoviruses have been recognized as unique plant viruses for many years but only since the early 1990's have they been classified as the only plant infecting members of the Bunyaviridae family. The type virus of the genus is tomato spotted wilt virus (TSWV) which remained little more than a research tool until the mid-1980's when several spotted-wilt like symptoms began showing up in other floral crops, peanuts, tobacco, and watermelon (Figs. 3, 4, and 5).
Some arthropod vectors of plant viruses are difficult to see and difficult to handle, but are efficient as vectors of viruses. Eriophyid mites have been implicated as the vectors of a new disease of corn and wheat observed in the 1990's in the high plains of Nebraska and other states (Figs. 7and 8). The disease resembled wheat spot mosaic and wheat spot chlorosis which were reported in the 1950's and 1960's but had never become widely important. The high plains virus (HPV) has now been reported from several countries outside the United States and may be a complex of viruses whose origins and interactions are not always identical but whose vector is the eriophyid mite
Its appearance is sporadic but losses may be catastrophic.
Diseases can be new and emerging or they can be threatening but not yet introduced or established in North America
Such is the case of Sharka, caused by plum pox potyvirus (PPV), one of the most serious diseases of
(peach, apricot, nectarine, plum, sweet and sour cherry, almond, and wild and ornamental types) (Figs. 9, 10, 11, and 12).

Figure 13.
Click for an enlarged view.

Figure 14.
Click for an enlarged view. |
This disease has been kept out of North America by very strict quarantine regulations. In 1994, Nemeth estimated that 100 million trees were infected in Europe. Sharka is found throughout Europe, Egypt, Turkey, Syria, India, and most recently Chile (1992) (Fig. 13). It was verified in the Santiago region in 1994 and now considered widespread in Chile. It is caused by several strains of PPV which are transmitted by several aphid species in a non-persistent manner (Fig. 14). It also is spread by propagative materials and possibly through seed .
Four strains of PPV have been identified in Europe and Egypt. The Marcus strain (PPV-M) is the most severe and is spread most readily, although the Dideron strain (PPV-D) is most common (found in Chile). Most recently PPV-C has been isolated and characterized from both sweet and sour cherry. All strains can be detected by graft inoculations to woody indicators P. tomentosa (IR473/1 X IR474/1) and P. persicae GF 305 seedlings, by monoclonal and polyclonal antisera, and several variations of RT-PCR. A paucity of resistance in Prunus has focused some effort toward genetically engineered resistance. |
An international collaboration between scientists at USDA-ARS, USDA-APHIS, and INRA (Bordeaux, France) has resulted in the transfer and expression of the PPV coat protein gene into plum and a highly resistant plum clone (C-5) has been identified (Fig. 15). This level of resistance has been transferred into plums through hybridization of transgenic plants to produce plum pox resistant plums.
 |
|
Figure 15. Plum pox strain D infecting susceptible transgenic plum clone C6; clone C5 highly resistant. |
Prunus crops are not the only fruit crops harboring serious viruses not found in the North American continent. Citrus production is unique as a woody perennial, which remains vegetative over many years and provides a good target for accumulation of viruses over time from surrounding plants or from new crops introduced into traditional citrus crop areas. Citrus species are affected by many different virus diseases, which form a spectrum from threatening to new and emerging because of a changing vector picture. With the establishment of the brown citrus aphid (Toxoptera citricida) in Florida in 1995, the threat of spread of severe stem pitting isolates of citrus tristeza virus has increased exponentially along with projected loss of an estimated 18 million trees from tristeza-induced decline. Virus and viruslike diseases of citrus are a dynamic system and new problems continue to arise. New problems may involve changes in vector dynamics associated with other crops or ingress of a pathogen from other hosts. The impact of these ingress events is determined by the potential for secondary spread, and the pathogen and vector reservoirs in other crops. Citrus chlorotic dwarf (CCD) (Figs. 16 and 17), a disease that appeared in citrus groves in Turkey after introduction of the bayberry whitefly (Parabemesia myricae), provides an illustration of a new citrus virus problem with a probable non-citrus origin, and a vector with citrus and non-citrus hosts. The bayberry whiteflies are established in Florida and other states which increases the risk of establishment if the pathogen is introduced.
 |
 |
| Figures 16 and 17. Click images for enlarged views. |
Whitefly Transmitted Crinivirus Diseases
Whiteflies, mentioned above as important vectors of geminiviruses, also have been implicated as vectors of a new group of viruses, the ‘Criniviruses,’ which were previously grouped with the closteroviruses. With the sudden explosion of whiteflies into more temperate regions in the last 10 to 20 years, several new diseases became important yield impacters. Lettuce infectious yellows (Fig. 18) alone caused losses of lettuce and cucurbits totaling multiple millions in a single season and eliminated fall melons in southern California and parts of Arizona. Another virus of this group, cucurbit yellow stunting disorder (Figs. 19 and 20), is the most important virus pathogen of cucurbits in southern Europe and the middle East and is a potentially threatening disease for North America.
 |
 |
 |
Figures 18, 19, and 20. Click any image for an enlarged view.
|
Members of the Criniviruses all induce yellowing symptoms in their plant hosts, are generally phloem-limited, non-mechanically transmissible, and have large ss-RNA genomes. Characterization of one of these viruses, lettuce infectious yellows virus (LIYV), showed that the LIYV genome was composed of two ss-RNAs of 8.1 and 7.2 kb (Fig. 21). RNA 1 encodes for proteins associated with RNA replication and RNA 2 encodes the hallmark closterovirus gene array. LIYV is now recognized as the type member of the genus Crinivirus within the family Closteroviridae. Are these new or emerging virus diseases caused by new virus isolates, are they renamed and reclassified because of improvements in molecular technology, or are they the result of a new vector biotype?
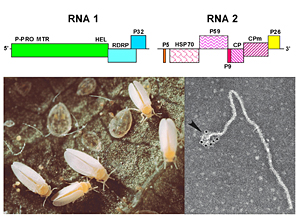 |
| Figure 21. Click for an enlarged view. |
Factors Contributing to an Increase in New and Emerging Viruses
There are many causes for the apparent increase in new and emerging viruses and in the knowledge gained of viruses not found on the North American continent. In almost all the above scenarios, man plays an important role. As misuse of antibiotics has led to the development of bacteria with multiple resistance, misplaced confidence in our ability to control vectors with pesticides has resulted in multiple chemical resistances (Fig. 22). Germplasm collecting expeditions, international movement of plant material and, lowered trade barriers offers opportunities for unknown viruses to be introduced into new areas. As we increase the use of transgenic crops carrying viral genes into new vegetative environments, will we continue to increase the rate of new virus evolution, or will we be able to control the viruses now present and create a stable, balanced ecological system?
 |
| Figure 22. Raspberry bushy dwarf idaeovirus. Click image for an enlarged view. |
Three components in the virus disease triangle, plant host, arthropod vector, and plant virus all play a role in the rise and fall in occurrence and spread of plant virus diseases. However, super-imposed over all these is the involvement of man as he manipulates the components and the environment in which they interact.
References
1. Acuna, R. 1993. Outbreaks of plum pox virus in Chile. Abstract, Conference on plum pox, Bordeaux, France.
2. Best, R. J. 1968. Tomato Spotted Wilt Virus. In: Advances inVirus Research 13:65-145. Ed. K. M. Smith, M. A. Lauffer. Academic Press, New York.
3. Candresse, T., MacQuaire, G., Lanneau, M., Quiot-Douine, L., Quiot, J. B., and Dunez, J. 1995. Analysis of plum pox virus variability and development of a strain-specific PCR assay. Acta Hortic. 386:357-369.
4. Çinar, A., Korkmaz, S., and Kersting, U. 1994. Presence of new whitefly-borne citrus disease of possible viral aetiology in Turkey. FAO Plant Prot. Bull. 42:73-75.
5. Daughtrey, M. L., Jones, R. K., Moyer, J. W., Daub, M. E., and Baker, J. R. 1997. Tospoviruses strike the greenhouse industry: INSV has become a major pathogen on flower crops. Plant Dis. 81:1220-1230.
6. De Avila, A. C., de Haan, P., Kormelink, R., Resende, De O., Goldbach, R. W., and Peters, D. 1993. Classification of tospoviruses based on phylogeny of nucleoprotein gene sequences. J. Gen. Virol. 74:153-159.
7. Duffus, J. E. 1987. Whitefly transmission of plant viruses. pp. 73-91. In: K. F. Harris (ed.). Current Topics in Vector Research. Springer-Verlag, New York.
8. Falk, B. W., and Bruening, G. 1994. Will growing transgenic crops generate new viruses and new diseases? Science 263:1395-1396.
9. Falk, B. W., Tian, T., and Yeh, H. -H. 1999. Luteovirus-associated viruses and sub-viral RNAs. Curr. Topics in Microbiol. and Immunol. 239:159-175.
10. Garnsey, S. M., Gottwald, T. R., and Yokomi, R. K. 1998. Control strategies for citrus tristeza virus. pp. 639-658. In: A. Hadidi, R. K. Khetarpal, and H. Koganezawa (eds.). Plant Virus Disease Control. APS Press, St. Paul
11. Hammond, J., Puhringer, H., da Camaro Machado, A., and da Camaro Machado, M. Laimer. 1998. A broad-spectrum PCR assay combined with RFLP analysis for detection and differentiation of plum pox virus isolates. Acta Hortic. 472:483-490.
12. Herrera, G., Sepulvida, P., and Madariaga, M. 1998. Survey of sharka disease (plum pox virus) on stone fruit trees in Chile. Acta Hortic. 472:393-399.
13. Klassen, V. A., Boeshore, M., Koonin, E. V., Tian, T., and Falk, B. W. 1994. Partial characterization of the lettuce infectious yellows virus genomic RNAs, identification of the coat protein gene and comparison of its amino acid sequence with those of other filamentous RNA plant viruses. J. Gen. Virol. 75:1525-1533.
14. Klassen, V. A., Boeshore, M., Koonin, E. V., Tian, T., and Falk, B. W. 1995. Genome structure and phylogenetic analysis of lettuce infectious yellows virus, a whitefly-transmitted closterovirus. Virology 208:99-110.
15. Korkmaz, S., Cinar, A., Kersting, U., and Garnsey, S. M. 1995. Citrus chlorotic dwarf: a new whitefly-transmitted viruslike disease of citrus in Turkey. Plant Dis. 79:1074.
16. Law, M. D., and Moyer, J. W. 1990. A tomato spotted wilt-like virus with a serologically distinct N protein. J. Gen. Virol. 71:933-938.
17. Law, M. D., Speck, J., and Moyer, J. W. 1992. The mRNA of Impatiens necrotic spot Tospovirus (Bunyaviridae) has an antisense genomic organization. Virology 188:732-741.
18. Levins, R., Albuquerque de Possas, C., Awerbuch, T., Brinkmann, U., Eckardt, I., Epstein, P., Makhoul, N., Puccia, C., Spielman, A., and Wilson, M. E. 1993. Preparing for new infectious diseases. Harvard School of Public Health. Boston, MA (Working Paper No. 8).
19. Levy, L., and Hadidi, A. 1994. A simple and rapid method for processing tissue infected with plum pox potyvirus for use with specific 3' non-coding region RT-PCR assays. EPPO Bull. 24:595-604.
20. Magome, H., Yoshikawa, N., Takahashi, T., Ito, T., and Miyakawa, T. 1997. Molecular variability of the genomes of capilloviruses from apple, Japanese pear, European pear, and citrus trees. Phytopathology 87:389-396.
21. Medina, V., Tian, T., Wierzchos, J., and Falk, B. W. 1998. Specific inclusion bodies are associated with replication of lettuce infectious yellows virus (LIYV) RNAs in Nicotiana benthamiana protoplasts. J. Gen. Virol. 78:2325-2329.
22. Moyer, J. W. (In Press - 1999). TOSPOVIRUSES (Bunyaviridae). In: Encyclopedia of Virology, eds. R. Webster and A Granoff. Academic Press, Ltd. London.
23. Nemeth, M. 1994. History and importance of plum pox in stone fruit production. EPPO Bull. 24:525-536.
24. Prins, M., and Goldbach, R. 1998. The emerging problem of tospovirus infection and nonconventional methods of control. Trends Microbiol. 6:31-35.
25. Qui, W. P., Geske, S. M., Hickey, C. M., and Moyer, J. W. 1998. Tomato spotted wilt tospovirus genome reassortment and genome segment-specific adaptation. Virology 244:186-194.
26. Ravelonandro, M., Scorza, R., Bachelier, J. C., Labonne, G., Levy, L., Damsteegt, V., Callahan, A. M., and Dunez, J. 1997. Resistance of transgenic Prunus domestica to plum pox virus infection. Plant Dis. 81:1231-1235.
27. Rocha-Pena, M. A., Lee, R. F., Lastra, R., Niblett, C. N., Ochoa-Corona, F. M., Garnsey, S. M., and Yokomi, R. K. 1995. Citrus tristeza virus and its aphid vector, Toxoptera citricida. Plant Dis. 79:437-445.
28. Rubio, L., Soong, J., Kao, J., and Falk, B. W. 1999. Geographic distribution and molecular variation of isolates of three whitefly-borne closteroviruses of cucurbits: lettuce infectious yellows virus (LIYV), cucurbit yellow stunting disorder virus (CYSDV) and beet pseudo-yellows virus (BPYV). Phytopathology (In Press).
29. Scorza, R., Callahan, A. M., Levy, L., Damsteegt, V., and Ravelonandro, M. 1998. Transferring potyvirus coat protein genes through hybridization of transgenic plants to produce plum pox resistance in plums (Prunus domestica L.). Acta Hortic. 472:421-428.
30. Scorza, R., Ravelonandro, M., Callahan, A. M., Cordts, J. M., Fuchs, M., Dunez, J., and Gonsalves, D. 1994. Transgenic plum (Prunus domestica) express the plum pox virus coat protein gene. Plant Cell Rep. 14:18-22.
31. Stahler, M. M., Lawrence, F. J., and Martin, R. R. 1995. Incidence of raspberry bushy dwarf virus in breeding plots of red raspberry. Hort. Science 30:113-114.
32. Tian, T., Klaassen, V. A., Soong, J., Wisler, G., Duffus, J. E., and Falk, B. W. 1996. Generation of cDNAs specific to lettuce infectious yellows closterovirus and other whitefly-transmitted viruses by using RT-PCR and degenerate oligonucleotide primers corresponding to the closterovirus gene encoding the heat shock protein 70 homolog. Phytopathology 86:1167-1173.
33. Tian, T., Rubio, L., Yeh, H. -H., Crawford, B., and Falk, B. W. 1999. Lettuce infectious yellows virus: in vitro acquisition analysis using partially purified virions and the whitefly Bemisia tabaci. J. Gen. Virol. 80:1111-1117.
34. Ullman, D. E., Sherwood, J. L., and German, T. G. 1997. Thrips as vectors of plant pathogens. pp. 539-565. In: T. L. Lewis, Thrips as Crop Pests, CAB International, London.
35. Wang, H., de A. Gurusinghe, P., and Falk, B. W. 1999. Systemic insecticides and plant age affect beet curly top virus transmission to selected host plants. Plant Dis. 88:351-355.
36. Wetzel, T., Candresse, T., Ravelonandro, M., and Dunez, J. 1991. A polymerase chain reaction adapted to plum pox virus detection. J. Virol. Methods 33:355-366.
