
Introduction

Fig. 1. Freshly hatched second stage (L2) larvae (Nomarski micrograph) (Meloidogyne incognita) (courtesy of Dr. David M. Bird, NC State University, Dept. of Plant Pathology). (Click image for larger view). | |
Plant-associated microbes play critical roles in agricultural and food safety and security and in the maintenance of ecosystem balance. Some of these diverse microbes, which include viruses, bacteria, oomycetes, fungi, and nematodes, cause plant diseases (Figs. 1 and 2) while others prevent diseases or enhance plant growth (Fig. 3). Despite their importance, as of June 2002, less than six percent of the microbes whose genomic sequence has been completed and made publicly available were plant-associated microbes (see The Institute for Genomic Research's comprehensive lists of
all genomes sequenced or in progress and
sequenced or in-progress phytopathogens, as well as the Department of Energy Joint Genome Institute's Program on Microbial Genomics). To remedy this serious deficiency in information, the microbe-plant interaction community, lead by the Public Policy Board of the American Phytopathological Society (APS), is developing a strategy to undertake the structural and functional genomic analysis of this diverse group of microbes that interact with plants. Here, we describe the rationale for an international public initiative for genomic analysis of plant-associated microbes.
|
 | |
 | |
|
Fig. 2. Tomato spotted wilt virus on peanut (courtesy of NC State University, Dept. of Plant Pathology). (Click image for larger view).
| |
Fig. 3. A soybean root infected with a vesicular arbuscular endomychorrhizal fungus (courtesy of NC State University, Dept. of Plant Pathology). (Click image for larger view). | |
Background
Microbes are found on or within every higher organism, including plants. Plant-associated microbes, which include viruses, bacteria, fungi, oomycetes, and nematodes, play critical roles in plant health. Some microbes cause diseases whereas others prevent diseases or enhance plant growth. Microbes also interact with each other on or within plants with the result of more or less severe disease. Within each microbe group, the diversity of species that interacts with plants is tremendous. For example, more than 10,000 pathogenic species of fungi and viruses are estimated to cause diseases on plants, and many of these are economically important (Fig. 4).
|

Fig. 4. Cedar-quince rust on hawthorn, caused by the fungal pathogen Gymnosporangium clavipes (courtesy of Kansas State University, Department of Plant Pathology). (Click image for larger view). | |
|

Fig. 5. Late blight (courtesy of Jean Ristaino). (Click image for larger view). |
Pathogenic microbes continually erode our supply of food and fiber, causing losses to producers of over $200 billion annually. Recent terrorist activities have heightened concern that, in the wrong hands, these agents could be used to threaten our food, environment, or economic security (see the
APSnet Feature on biosecurity). The historic devastation wrought by the inadvertent introduction of exotic pathogens testifies to such potential. For example, the introduction of the chestnut blight pathogen,
Cryphonectria parasitica, from Europe in 1904 effectively eliminated from North America the American chestnut, an important and beautiful timber tree. Similar potential damage is currently occurring with disease epidemics of forest trees including sudden oak death caused by
Phytophthora ramorum and pitch canker of pine caused by
Fusarium circinatum. Introductions of other pathogenic microbes have caused tremendous suffering to humans, including the famines that resulted from the
Phytophthora late blight (Fig. 5) epidemic of potato in 1845-1846 in Ireland and the
Cochliobolus brown spot epidemic of rice in Bengal in 1943.
Despite their importance to our agricultural base and food security, we know little of the molecular genetics of these microbes. Our understanding of how these microbes benefit plants or cause disease is rudimentary at best. With the recent advent of genomics, which includes the structural and functional analysis of the microbe genes and the proteins encoded by those genes, we see the opportunity to understand the nature of beneficial and pathogenic microbe-plant-associations at levels never before possible.
The Promise of Genome Analysis of Plant-Associated Microbes
The knowledge of how microbes interact with plants is essential to the development of effective, environmentally sound, chemically-based strategies for disease control. In the US, over $600 million are spent annually on agricultural fungicides alone. Similar to what is being observed with the heavy use of antibiotics targeted to human or animal pathogens, changes in pathogen biology more and more frequently render some of these chemicals ineffective. Furthermore, increased regulatory policies are restricting the use of existing agrochemicals for pathogen control. With genome information, multiple tactics for control of pathogens could be developed. For example, the identification of more precise targets in the pathogen may allow for design of more specific and effective chemicals that are environmentally benign.
The powerful automated sequence technologies currently available and the advances in bioinformatics have made the task of sequencing entire genomes of organisms almost routine. The sequenced genomes of a few model organisms are already enabling a wide variety of new discoveries, including new genes and metabolic pathways and insights into the mechanisms of microbial pathogenesis. The continuous improvement of bioinformatics tools is enhancing the discovery power of these sequences. With these exciting advances, the means are now available to generate sequence and function information, as are the tools to use that information to understand the basic biology of microorganisms that cause or prevent diseases on plants.
By enhancing our knowledge base, genome analyses will provide tools to abrogate the problems caused by plant pathogenic microbes through genetically-based approaches and will allow development of improved beneficial microbes. Historically, we have studied the molecular basis for interactions between plants and microbes using a gene-by-gene approach and have used host plant resistance as a major control approach. Now, structural and functional genomic analyses will increase the speed of identification of genes involved in host-pathogen interactions and will allow genome-wide approaches to understanding the role of a gene or pathway in interactions with plants. Some genes will potentially be useful as sources of pathogen-derived resistance, as has already been demonstrated for many viral diseases. Other genes may be involved in activating plant defense responses.
Comparisons of genomes of related strains or species will provide an understanding of the evolution of microbial genomes, particularly as they evolve in associations with plants. Researchers exploiting comparative genomics will be able to predict the molecular basis of how some microbes have evolved to form intimate biotrophic associations with plant cells, whereas others inhabit intercellular spaces and still others colonize only the vascular systems of their hosts. Comparisons of sequences within and between species will also provide information to develop accurate diagnostic tools. Such precision in diagnostics may be essential to track the source or origin of a pathogen (e.g., as for the anthrax strains recently released in the US or plum pox virus recently introduced). This information may be essential for forensic purposes as well as to understand the genetic potential of the pathogen to become established or to cause an epidemic. Additionally, because of the evolutionary conservation between microbes of different genera and because of the exchange of genetic information between some groups of microbes, genome analysis of plant-associated microbes will contribute to our understanding of how animal pathogens cause disease. For example, bacterial genes involved in plant disease induction exhibit a great deal of similarity to bacterial genes involved in disease induction in animals and humans (see the Proceedings of the National Academy of Sciences paper,
Pathogens and Hosts: The Dance is the Same, the Couples are Different). Thus, identifying the full complement of genes involved in pathogenesis will help decipher the disease strategies of these organisms and will guide design of new control measures for use in both plants and animals.
|
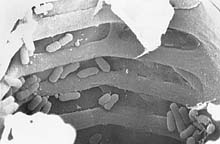
Fig. 6. Scanning electron micrograph showing cells of the rice bacterial blight pathogen,
Xanthomonas oryzae pv.
oryzae, in rice xylem vessels. (Click image for larger view). | |
In sum, functional, structural and comparative genome analyses will provide insights into how microbes infect and establish either disease or beneficial interactions with host plants, how they reproduce and spread in the plants, and how they persist and become disseminated in the environment. This information will be critical for the development of more effective, less harmful control measures and of accurate diagnostic and tracking methods.
Which Plant-Associated Microbes Should Be Sequenced?
Even with the costs of genome sequencing decreasing, it is clearly not feasible to sequence genomes of all plant-associated species at this time. Due to the wide variety of organisms that cause or prevent plant diseases, the APS members elected to focus possible sequencing efforts for maximal benefit. Through a series of task forces with broad input, the APS established a prioritized list of species for genome sequencing. This list, entitled
“Microbial Genomic Sequencing: Perspectives of the American Phytopathological Society,” was developed using the following criteria:
· Economic importance and relevance to U.S. and world-wide agriculture;
· Unique biological or environmental features;
· Broad interest to a significant community of scientists or agriculturists;
· Genetic tractability (i.e., the ease with which genetic studies, such as crosses, genome modifications, etc., can be performed); and
· Availability of tools and other biological resources (e.g., gene libraries, genetic maps, and genetic tractability of the host so that the system could be addressed from both sides of the host-pathogen interaction, etc.).
The APS list, compiled in 2000-2001, was intended to be dynamic, consistent with the inconstant nature of plant health problems, and would be subject to change. To encourage even wider input and a more accurate up-to-date list, the research community has been invited to revisit the list criteria and content, and make further recommendations through the chairs of each pertinent subject matter committee. These comments/recommendations will be the subject of a discussion forum at the APS meetings in July 2002. In addition, the need for specialized lists, such as the list of organisms that are potential introduced threats to U.S. agriculture (see the
APSnet Feature on nonindigenous plant pathogens threatening U.S. crops and forests), will be discussed.
|

Fig. 7. Blast (gray leaf spot) on rye grass caused by Magnaporthe grisea, which is currently being sequenced (photo courtesy of Kansas State University, Department of Plant Pathology).
| |
Community Discussions Towards a Plant-Associated Microbe Genome Initiative
A workshop was held in Washington, D.C., April 9-11, 2002 to develop a strategy to expedite genome analysis of plant-associated microbes. The workshop was organized by the APS Public Policy Board, and was funded by the Department of Energy (DOE), the National Science Foundation (NSF), USDA-Agricultural Research Service (USDA-ARS), and USDA-National Research Initiative (USDA-NRI). Participants included academic, industrial, and governmental experts from the genomics and microbial research communities and observers from the federal funding agencies. The participants agreed that:
· the lack of publicly available information on the genomes of plant-associated microbes is a major impediment to research, and, as a result, to crop biosecurity;
· the APS list of microbes for sequencing is a good starting point, but requires periodic review and updating;
· current technological and biological resources would allow for sequence analysis of a significant number of microbes from this very diverse group, given appropriate funding resources. The availability of genome sequence would enable and enhance the development of tools for functional and comparative genomics; and
· there is an urgent need for development and employment of flexible and broadly applicable bioinformatic tools and the establishment and continual updating of databases and genetic resource centers.
An advisory committee was appointed to oversee the development of a Plant-Associated Microbe Genome Initiative. Discussions toward this initiative will be broadened by (1) publication of a draft position paper via society newsletters and email lists, inviting the research community to contribute ideas, and feedback through email (microbegenomics@scisoc.org); and (2) through a public open forum held at the Annual APS meetings in Milwaukee, WI (July, 2002). At this forum, the Plant-Associated Microbe Genome Initiative will be summarized and feedback will be solicited. All comments and suggestions from these various sources will be used to develop a final position paper that will be circulated among the federal agencies that are interested in promoting genome analysis of these important microbes.
Recommendations for Genome Analysis of Plant-Associated Microbes
To remedy the paucity of genomic information available for plant-associated microbes, a five-year concentrated effort, called a Plant-Associated Microbe Genome Initiative is proposed. The initiative reflects the diversity of the microbes that interact with plants and the tremendous differences in the current status of the genome analysis of those organisms. Due to the diverse group of organisms, the initiative provides a broad outline of how an international comprehensive effort might be approached and does not provide specific technical detail of how the analyses should be accomplished. To be effective, the effort must be
collaborative, well-coordinated, and
international. Industrial and public partnerships should be encouraged, with the caveat that all information must be made rapidly available to the
public. Finally, the effort must involve strong
education/training programs.
What Research Areas Should Be Targeted?
Structural analysis. Structural sequence information is the key to gene discovery and the first step towards learning the function of each gene. There are vast differences in the amounts of sequence information currently available for different plant-associated microbes. For example, due to their smaller genome size, many different plant viruses and a few plant bacteria have already been sequenced, whereas no genomes of plant-associated oomycetes, fungi, or nematodes have been sequenced to completion. These differences in available information, which vary widely within and between microbe groups, impact the entry level for analysis of each group. They also impact the cost of sequencing the genomes of different microbe groups.
|

Fig. 8. Soybean cyst nematode (Heterodera glycines) (courtesy of NC State University, Dept. of Plant Pathology). (Click image for larger view).
| |

Fig. 9. Root knot nematode on carrot (Meloidogyne spp.) (courtesy of NC State University, Dept. of Plant Pathology). (Click image for larger view).
| |
Although discussed extensively, no consensus was reached on the depth of sequence needed (i.e., draft versus complete genome sequences). One component of workshop participants suggested that to obtain useful SNP and genotype information more than one strain of an organism should be sequenced. As an example, this component recommended that the genomes of three strains of a given bacterial species should be sequenced completely followed by sequencing to 8X coverage of an additional 10 to 20 strains. Another component suggested that if a complete genome sequence were available from one type strain, then lower coverage draft sequence of additional strains would provide information sufficient for most purposes. Although there were other permutations of this discussion, the emerging conclusions are that the depth of sequencing will depend on the end user needs. For example, the depth of the sequence could depend on the questions relevant to that organism, and this relevance will likely vary with the complexity of the microbial genome of interest and available resources.
Recommendations for the Plant-Associated Microbe Genome Initiative:
· Focus initial resources largely on sequence analysis to remedy the paucity in sequence information available for plant-associated microbes;
· Target 2,000 Mb of sequence total over all microbes over five years; and
· Allow research community to direct the depth of sequence or coverage for each microbe, based on specific needs or questions.
Functional analysis. Sophisticated technologies, including array-based technologies, are available for both gene expression and proteomic analyses and will enable the dissection of the functions of genes and proteins involved in interactions of these microbes with plants and the environment. Additionally, high-throughput genome-wide gene deletion and tagging methods are being developed. Although limited to a few groups of microbes, there is publicly-available genome sequence information currently for a few plant-associated microbes that is sufficient to launch functional genomic studies. These microbes can serve as model systems for functional genomic efforts to identify genes/proteins and biochemical pathways that are involved in interactions with plants.
|

Fig. 10. Microarray technology will be useful for functional genomics and for diagnostics. (Click image for larger view). | |
Recommendations for the Plant-Associated Microbe Genome Initiative:
· Focus initial resources on those few microbes with sufficient structural genomic and genetic resources available; and
· Invest in improvement of technologies for functional analyses.
Standardized bioinformatic tools and database system. The underpinning for successful structural and functional genome analysis is the ability to integrate, manipulate, analyze, compare, and store all of data generated by these technologies. It was clear from the intensity of discussions surrounding the bioinformatic tools and database systems that this is an area of major deficiency and concern. It was also clear that this is not just a problem for analysis and storage of data for the genomes of plant-associated microbes. Databases are a particular concern. Participants discussed the merits of fixing the existing systems as opposed to developing new, state-of-the-art systems, or, using one major database as opposed to many smaller, interconnected databases. Critical features highlighted were that these tools must be (1) flexible (e.g., usable for comparisons among different genomes, including microbes and plants), (2) scalable, (3) inter-operable, (4) easily implemented, (5) readily updateable, and (6) stably funded and maintained. Guidelines should be developed for standardization of public databases, and these should include common (1) means for electronic submission of data, (2) terminology, and (3) criteria for quality of data submitted.
Recommendations for the Plant-Associated Microbe Genome Initiative:
· Increase resources allocated to the standardization of databases and bioinformatic tools, as these are essential to the total science of genomics and the application of that science;
· Develop mechanisms for discussions, planning, and research at an international level; and
· Develop a framework to finance and manage the maintenance and continual updating of databases and information resources.
Training the next generation of plant microbiologists. To capitalize on the information provided from the analysis of the genomes of plant-associated microbes, a key goal of the
Plant-Associated Microbe Genome Initiative must be to both train new and update current plant microbiologists such that in the future, we have a broadly trained research community with skills in microbe and plant biology and in computational sciences. Additionally, these scholars will need the skills to interact and collaborate in an international research arena.
Recommendation for the Plant-Associated Microbe Genome Initiative:
· Train plant microbiologists in the skills of genomics and computational sciences (e.g., this training should involve scientists at all levels of their careers to ensure an up-to-date, skilled pool of plant microbiologists); and
· Provide funding and opportunities for domestic and international research collaborations.
Additional Resources
Sequenced genomes list from The Institute for Genomic Research (TIGR)
Sequenced phytopathogens list from The Institute for Genomic Research (TIGR)
Program on Microbial Genomics of DOE's Joint Genome Institute
"Crop Biosecurity," an APSnet Feature
"Pathogens and Hosts: The Dance is the Same, the Couples are Different," from the Proceedings of the National Academy of Sciences
Microbial Genomic Sequencing: Perspectives of the American Phytopathological Society
"What are the Nonindigenous Plant Pathogens that Threaten U.S. Crops and Forests?," an APSnet Feature
