Introduction
Many plant pathogenic fungi utilize toxins for successful pathogenesis of their hosts. Among these are the photoactivated perylenequinones, produced by a number of important fungal plant pathogens, including species of Alternaria, Cercospora, Cladosporium, Elsinoe, and Hypocrella, among others. The toxins produced by these pathogens are red in color and share a similar structure (Fig. 1). Of the perylenequinone toxins, the best studied are the hypocrellins and cercosporin (8,11). The hypocrellins have been studied, not for their role in plant disease, but rather due to their pharmaceutical potential as anticancer agents through photodynamic tumor therapy. Cercosporin, by contrast, has been shown to play an important role in Cercospora diseases of diverse hosts. Here we discuss our long-term studies on the biosynthesis and toxicity of cercosporin and our efforts to utilize this understanding for the development of Cercospora-resistant crops.
| |
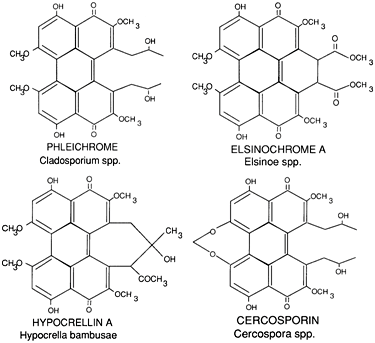
Fig. 1. (A) Structure of perylenequinone toxins produced by plant pathogenic fungi. |
|
| |
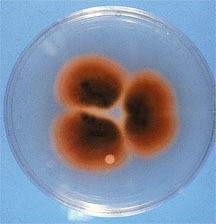
Fig. 1. (B) Culture of Cercospora nicotianae showing red pigmentation on underside of colony due to production of cercosporin. |
|
Cercosporin Mode of Action
Cercosporin and the other perylenequinone toxins are photoactivated and lack toxicity in the dark. In the light, these compounds absorb light energy and are converted to an energetically activated triplet state. The triplet molecule then reacts with oxygen and results in the generation of toxic, activated oxygen species such as singlet oxygen (¹O2) and superoxide (O2-.) (Fig. 2). This property classifies cercosporin as a photosensitizer, a term that dates to the early 20th century and was used to describe compounds that "sensitized" cells to visible wavelengths of light (18). Photosensitizers are structurally diverse, and include common dyes such as methylene blue and acridine orange as well as natural products such as porphyrins, flavins, chlorophyll, and extended quinones. Toxicity of photosensitizers is due to oxidative damage to lipids, proteins, and nucleic acids, with the cellular target dependent on where the photosensitizer molecule localizes in cells (22). Cercosporin is a membrane sensitizer and a potent producer of singlet oxygen. Exposure of plant cells and tissues to cercosporin results in peroxidation of the membrane lipids, leading to membrane breakdown and death of the cells (6). We hypothesize that membrane damage allows for leakage of nutrients into the leaf intercellular spaces, allowing for fungal growth and sporulation (Fig. 2).
| |
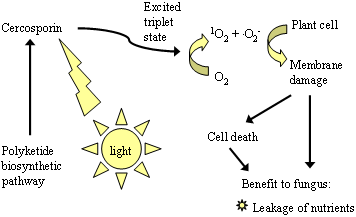
Fig. 2. The mode of action of cercosporin. Cercosporin is activated by light and, in the activated state, reacts with oxygen to produce toxic oxygen species such as singlet oxygen (¹O2) and superoxide (O2-). Production of reactive oxygen species leads to peroxidation of lipids in the plant cell membranes. It is hypothesized that this membrane damage allows for leakage into the intercellular spaces of nutrients that are used by the fungus to colonize the leaf. |
|
Biosynthesis of Cercosporin
Compared to the progress made on understanding cercosporin toxicity, an understanding of the cercosporin biosynthetic pathway and its regulation is only just beginning. The first report of cercosporin biosynthesis was made by a Japanese research group in the 1970s. Okubo and co-workers (16) analyzed culture filtrates after feeding Cercospora kikuchii with radioisotope-labeled acetate. They concluded that cercosporin synthesis proceeds via a fungal polyketide pathway, likely starting with a condensation of acetate and malonate molecules. Unfortunately, no further experiments followed. Since then, no chemical intermediates or enzymes directly involved in the cercosporin biosynthetic pathway have been identified.
Considerable work has been done on the regulation of cercosporin production. Light is the most critical factor, not only for toxicity, but also for cercosporin production (13). In both cases, visible light of the wavelengths absorbed by the cercosporin molecule (approximately 400-600 nm) are responsible for both inducing production and causing toxicity. Cercosporin production is completely suppressed under complete darkness, and brief exposure to light immediately triggers its biosynthesis. Other physiological parameters, including temperature, nutrient conditions, pH, source of carbon or nitrogen, and carbon:nitrogen ratio, all affect cercosporin production (7). Cercosporin production is inhibited at high temperatures (30°C; 86°F). Cercosporin is synthesized only in vegetative cultures, and is repressed under nutrient conditions that induce conidiation, such as growth on V8 juice medium. With C. nicotianae, we found that a thin layer of Difco potato dextrose agar (PDA) medium (less than 15 ml of medium) in a Petri dish supports the highest production of cercosporin under light (2). Other brands of PDA, PDA made with fresh potatoes, or other synthetic media drastically reduces cercosporin production under light. Further, production of cercosporin in culture is highly variable among species and even among isolates of the same species.
Through selection of the genes that are highly induced by light, Upchurch and co-workers identified the first gene, named CFP (cercosporin facilitator protein), required for cercosporin accumulation (1). The CFP protein has a strong similarity to the major facilitator superfamily (MFS) of membrane transporters. Deletion of the CFP gene in C. kikuchii created a mutant partially defective in both cercosporin production and resistance. Thus, CFP presumably is required for export of cercosporin out of the mycelium.
Cercosporin is a red pigment, and toxin-deficient mutants can be visually identified on agar plates. To identify cercosporin biosynthetic genes, we adapted a strategy using restriction enzyme-mediated insertion (REMI) mutagenesis and screened for toxin-deficient mutants in C. nicotianae (5). Sequences recovered from a REMI mutant completely defective in cercosporin production showed a strong similarity to many fungal polyketide synthases (PKSs). Expression of the gene, named CTB1 (cercosporin toxin biosynthesis), was highly regulated by light and medium composition, and regulation was correlated with cercosporin- producing conditions (2). Further analysis using a chromosome walking strategy has recently allowed us to identify a core CTB gene cluster, comprised of eight genes required for cercosporin biosynthesis.
We are characterizing the genes in this cluster, and have developed a working model for the biosynthetic pathway (Fig. 3). The polyketide synthase gene, CTB1, encodes a protein of 2196 amino acids containing conserved sequences: a keto synthase (KS), an acyltransferase (AT), a thioesterase (TE), and two acyl carrier protein (ACP) domains (2). Our research to date has identified some of the steps in the pathway and genes encoding some of the pathway enzymes (Fig. 3). Similar to fatty acid biosynthesis, the functional domains in CTB1 are responsible for condensation of acetyl-CoA and malonyl-CoA units (step 1), chain elongation to form a pentaketide molecule (step 2), and ring closure (step 3) in the early steps of cercosporin biosynthesis. During ring closure, oxidation and hydration reactions take place to form aromatic rings. The oxidation reactions are likely completed by a monooxygenase domain encoded by CTB3 (9), followed by hydration steps. Once the ring is closed, methylation steps (step 4) are operated by methyltransferases domains encoded by CTB3, resulting in polyketomethylene units. Cercosporin has a bilateral symmetric structure; thus, cercosporin is likely formed by fusing two identical polyketomethylene units (step 5, dimerization). This step can likely be completed enzymatically or nonenzymatically. Once it is synthesized, cercosporin must be exported out of fungal cells, presumably by the function of a major facilitator superfamily (MFS) transporter encoded by CTB4 (unpublished results). CFP, the MFS transporter that has previously been shown to be required for cercosporin production, is not found in the cluster, but likely also plays a role in transport.
| |
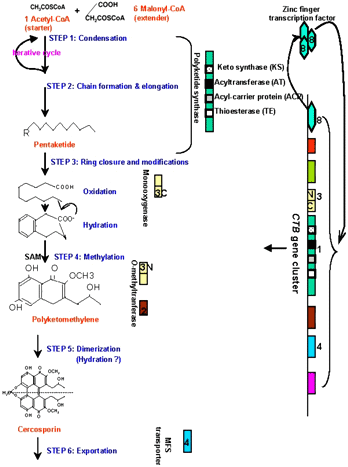
Fig. 3. Schematic illustration of a proposed cercosporin biosynthetic pathway. The putative functions of the CTB gene products in relation to cercosporin production are indicated (see text for details). |
|
Recent studies have shown that expression of the CTB genes identified to date correlate with environmental conditions that regulate cercosporin production, and that this regulation was controlled through a transcription factor (CTB8) (unpublished results). Other studies suggest that production of cercosporin is mediated by multiple signal transduction pathways. Studies using pharmacological inhibitors implicated the involvement of calcium and calmodulin signaling in cercosporin production (3). A study also revealed that cercosporin production was associated with a gene encoding a MAP kinase kinase kinase in C. zeae-maydis (17). In addition, cercosporin biosynthesis is regulated by another transcription factor, CRG1. The CRG1 gene was originally identified as a gene involved in cercosporin resistance, but disruption mutants also showed a reduction in production.
Combining the information known to date about the biosynthetic cluster and the environmental cues and genes involved in regulation, a model addressing the regulation of cercosporin biosynthesis is proposed (Fig. 4). Light is the primary signaling cue to activate cercosporin biosynthesis, through a complicated network involved Ca2+/Calmodulin and MAP kinase pathways (3,17). The zinc cluster transcription factor, CRG1, has been shown to regulate genes in the CTB gene cluster, including the regulator CTB8. Once CTB8 is produced, the entire biosynthetic pathway is induced. We have also observed possible feedback regulation caused by disruption of genes in the cluster (unpublished results). Studies are continuing to fully characterize the CTB genes and pathway intermediates, which will lead to a fuller understanding of the pathway for this important polyketide toxin and for pathway regulation.
| |
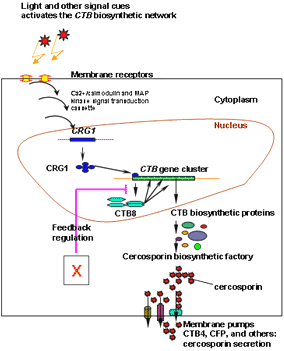
Fig. 4. Proposed model of signal transduction and regulatory controls for cercosporin biosynthesis and self-protection in Cercospora nicotianae. Light activates cercosporin biosynthesis through a network involved Ca2+/Calmodulin and MAP kinase pathways. The zinc cluster transcription factor, CRG1 regulates cluster genes including the specific pathway regulator CTB8. CTB8 induces the pathway. |
|
Role of Cercosporin in Disease
Several approaches have been used to study the importance of cercosporin in disease development (7) for the Cercospora species that synthesize this toxin. Cercosporin can be isolated from lesions on infected plants, indicating its production during infection. Ultrastructural studies of C. beticola-infected sugar beet documented membrane damage as a primary symptom, consistent with the mode of action of cercosporin. Studies in several Cercospora-host systems have documented the importance of light in normal symptom development. For example, shading has been shown to reduce penetration of coffee leaves by C. coffeicola, leading to a reduction in the number of lesions. On sugar beet, symptoms of C. beticola infection were delayed and attenuated when plants were grown under low light. Unlike the study with coffee, no reduction was noted in penetration, suggesting that colonization within the leaf is also impacted by low light conditions.
Further analysis of the importance of cercosporin in disease development has been done with mutants of several species that are reduced or deficient in cercosporin biosynthesis (8). C. kikuchii mutants disrupted for the CFP transporter described above had significantly reduced symptom development on soybean. Disruption of a MAP kinase gene (CZK3 MAPKKK) in C. zeae-maydis mutants interfered with both cercosporin production and conidiation, and also led to fewer lesions and chlorotic rather than necrotic symptoms on corn (17). In our work, we have disrupted the gene (CTB1) encoding the polyketide synthase responsible for cercosporin production in C. nicotianae (2). These mutants produce no cercosporin (Fig. 5A). When inoculated onto tobacco, the ctb1 mutants produced fewer lesions and did not cause the necrotic blighting symptom characteristic of wild type infection (Fig. 5B). These results demonstrate that, although not absolutely required for infection, cercosporin is a virulence factor that significantly enhances the amount and severity of disease.
 |
|
 |
|
Fig. 5. (A). TLC plate showing production of cercosporin by the C. nicotianae wild type (WT), but no production by mutants disrupted for the cercosporin polyketide synthase (CTB1) (T1 – T32). |
|
Fig. 5. (B). Symptom expression on tobacco caused by the wild type C. nicotianae (left) and ctb1 disruption mutant (right). Figures from Choquer et al. 2005. Mol. Plant-Microbe Interact. 18:468. |
Autodefense by Cercospora Fungi Against Cercosporin
Given the evidence for an important role for cercosporin in disease severity, we hypothesized that defense mechanisms targeted against cercosporin may provide an additional strategy for developing resistance to these important pathogens. Due to its production of activated oxygen species, cercosporin shows almost universal toxicity to cells, with activity documented against not only plants, but also bacteria, most fungi, and mice (8). The only organisms that show high levels of resistance to cercosporin are Cercospora species and other fungi that produce perylenequinone toxins, such as Alternaria and Cladosporium. Although levels as low as 1µM cercosporin kill plant cells, cultures of Cercospora species produce mM levels of cercosporin in culture and are unaffected. Thus our labs and others have studied the mechanisms that Cercospora fungi use for autoresistance to cercosporin, with the goal of using fungal resistance genes as a novel source of resistance in transgenic plants.
Several approaches have been used to identify genes for resistance. Upchurch and co-workers characterized genes in C. kikuchii that were specifically induced by light, as light is required for both production and activity of cercosporin. Among the genes identified by that group was CFP described above. Not only was this gene shown to encode a transporter involved in cercosporin production, but cfp disruption mutants were also shown to be more sensitive to cercosporin than was the wild type strain. Further, expression of CFP in a cercosporin-sensitive fungus (Cochliobolus heterostrophus) enhanced resistance to cercosporin (20), presumably by transporting cercosporin out of the cell.
Increased cercosporin resistance in bacteria has been identified in strains of Xanthomonas campestris pathovar zinniae that were identified by their ability to degrade cercosporin into a non-toxic compound, xanosporic acid (19). Cercosporin degradation was shown to require a gene encoding a putative oxidoreductase. Five independent non-degrading mutants all had mutations in the oxidoreductase gene, and degradation ability was restored when the mutants were transformed to express the wild type gene. This gene was also significantly up-regulated when the bacteria were exposed to cercosporin.
Other cercosporin resistance genes have been identified through screening of C. nicotianae mutants that are sensitive to cercosporin (8). Genes required for resistance were then identified by complementing the mutants with a library from the wild type strain. This approach identified three genes as essential for cercosporin resistance. Two of these were shown to encode enzymes in the vitamin B6 pathway and the third gene encodes a zinc cluster transcription factor.
The discovery of the vitamin B6 biosynthetic genes as essential for cercosporin resistance was an interesting discovery, as this vitamin, unlike other vitamins such as vitamin E and vitamin C, had not been known to play a role in antioxidant defenses. Vitamin B6 (pyridoxal phosphate and its vitamers) is an essential co-factor for enzymatic reactions, most notably transamination reactions involved in amino acid synthesis. We have subsequently shown, however, that B6 vitamers quench singlet oxygen and superoxide and have antioxidant activity (10,12), providing an explanation for its cercosporin-protecting activity.
Finally, the CRG1 transcription factor described above was shown to affect cercosporin resistance as well as production (3). Mutants disrupted for crg1 are highly sensitive to cercosporin, along with being suppressed for production. Thus, we hypothesize that CRG1 regulates cercosporin-resistance genes. In order to identify the genes regulated by CRG1, we created a subtractive library containing sequences that are differentially regulated between the crg1 mutant and wild type. This library contains 206 unique sequences that are currently being characterized for a possible role in cercosporin resistance.
Strategies for Engineering Resistant Plants
Several studies have tested the efficacy of the microbial genes described above in providing resistance when transformed into plants. Unfortunately, thus far these studies have been met with limited or no success. The most effective approach to date has been with the cercosporin transporter, CFP. Expression of the C. kikuchii CFP in tobacco did not reduce the number of lesions following inoculation with C. nicotianae; however, the size of lesions was reduced (21), suggesting that CFP may be a useful gene for engineering for resistance.
Other efforts have not been as successful. The X. campestris pv. zinniae oxidoreductase gene, although absolutely required for cercosporin resistance, proved not to be sufficient on its own to impart cercosporin-degradation activity, either to non-degrading bacteria (19) or to tobacco callus (unpublished results). Extensive efforts to identify additional genes involved in cercosporin degradation through selection and complementation of non-degrading X. campestris pv. zinniae mutants has thus far been unsuccessful. We hypothesize that such mutants may be lethal, and that an alternate approach, such as searching for cercosporin-regulated genes, may allow for the identification of other genes essential for degradation and resistance.
Considerable efforts were directed toward engineering tobacco for constitutive expression of the vitamin B6 genes, with a goal of enhancing not only cercosporin resistance, but also as a model to engineer increased abiotic stress resistance as well as improved nutritional content of crop plants. Tobacco was transformed to express the C. nicotianae PDX1 and PDX2 genes, which encode the major enzymes required for vitamin B6 synthesis (15). We co-transformed haploid tobacco with two plasmids containing the individual genes. PDX1 was expressed under the control of the CaMV 35S promoter with a TMV translational enhancer, and was cloned in a vector with kanamycin selection; PDX2 was driven by the A. tumefaciens ocs-mas chimeric promoter in a construct that allowed for selection with bialaphos, an herbicidal compound. Transformed plants were selected in vitro for kanamycin resistance, and further screened in the greenhouse by spraying with bialaphos. Plants with resistance to both markers were put through a chromosome doubling protocol, resulting in the recovery of diploid plants homozygous for both PDX genes (Fig. 6).
| |
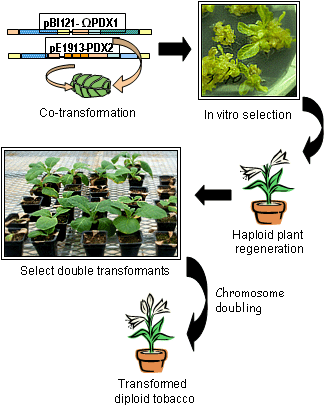
Fig. 6. Transformation of tobacco to express the Cercospora nicotianae vitamin B6 biosynthesis genes, PDX1 and PDX2. Haploid tobacco was co-transformed with PDX1 and PDX2 on plasmids containing selectable markers for kanamycin and bialaphos resistance, respectively. Transformed plants were selected in vitro for kanamycin resistance and further screened in the greenhouse by spraying with bialaphos. Plants with resistance to both markers were put through a chromosome doubling protocol, resulting in the recovery of diploid plants homozygous for both PDX genes. |
|
Although stable transformed lines were recovered that expressed both genes, only one line had a statistically significant increase in levels of the B6 vitamers, and the increase was small (approximately 15%). This line was the only line that also showed increased activity of the PDX1 enzyme. Subsequent molecular analysis indicated that transformation with the fungal PDX genes resulted in down-regulation of the endogenous tobacco PDX genes, suggesting that the pathway is tightly regulated. The line with elevated B6 levels differed from the other lines in having high expression of the transgenes. We tested seedlings of this line for resistance to cercosporin toxicity, but no differences were found, indicating that a modest increase in B6 levels is not sufficient to impart cercosporin resistance. As a comparison, C. nicotianae was found to have 2-3 fold higher B6 levels than the cercosporin-sensitive fungi Neurospora crassa and Aspergillus flavus (15), suggesting that major increases in B6 levels would be needed to provide resistance. Although other strategies may allow for increasing B6 levels in plants, our results suggested that increased B6 levels may be debilitating to plants, resulting in delayed germination and slow seedling growth. Studies in yeast have also documented growth impairments in strains transformed to express B6 genes. Thus a strategy of increasing B6 levels may hold promise for providing cercosporin resistance, but may have unacceptable side effects for the plant.
Our current efforts are focused on genes recovered from the subtractive library described above. Among these are several genes that encode putative MFS and ATP binding cassette (ABC) transporters. The role that the CFP transporter plays in cercosporin resistance was described above. In addition, studies in yeast and Botrytis have also identified ABC and MFS transporters that provided resistance to cercosporin (14,23). We are currently characterizing the role of our subtractive library transporters in cercosporin resistance and production using gene disruption techniques.
Conclusion
Cercosporin plays an important role in Cercospora pathogenicity and lesion formation in several crop species. Thus, any means of reducing cercosporin toxicity promises to have an important application for managing Cercospora leaf spot diseases in the field. In addition, cercosporin is very toxic due to the production of reactive oxygen species. Breeding for Cercospora disease resistance has been difficult since cercosporin has general toxicity to cells. Through our studies, we found that biosynthesis and regulation of cercosporin is a complicated process. Also, the mechanisms by which the Cercospora fungi operate to protect themselves against cercosporin toxicity likely involve multiple factors. Continued research in understanding these cellular defense mechanisms and in developing strategies for blocking cercosporin production will lead to better management of Cercospora diseases in the future.
Literature Cited
1. Callahan, T., Rose, M., Meade, M., Ehrenshaft, M., and Upchurch, R. 1999. CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Mol. Plant-Microbe Interact. 12:901-910.
2. Choquer, M., Lahey, K. A., Chen, H.-L., Cao, L., Ueng, P. P., Daub, M. E., and Chung, K. R. 2005. The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin toxin biosynthesis and fungal virulence in Cercospora nicotianae. Mol. Plant-Microbe Interact. 18:468-476.
3. Chung, K. R. 2003. Involvement of calcium/calmodulin signaling in cercosporin toxin biosynthesis by Cercospora nicotianae. Appl. Environ. Microbiol. 69:1187-1196.
4. Chung, K. R., Daub, M. E., Kuchler, K., and Schuller, C. 2003. The CRG1 gene required for resistance to the singlet oxygen-generating cercosporin toxin in Cercospora nicotianae encodes a putative fungal transcription factor. Biochem. Biophys. Res. Commun. 302:302-310.
5. Chung, K. R., Ehrenshaft, M., Wetzel, D. K., and Daub, M. E. 2003. Cercosporin-deficient mutants by plasmid tagging in the asexual fungus Cercospora nicotianae. Mol. Gen. Genet. 270:103-113.
6. Daub, M. E., and Briggs, S. P. 1983. Changes in tobacco cell membrane composition and structure caused by the fungal toxin, cercosporin. Plant Physiol. 71:763-766.
7. Daub, M. E., and Ehrenshaft, M. 2000. The photoactivated Cercospora toxin cercosporin: Contributions to plant disease and fundamental biology. Ann. Rev. Phytopath. 38:461-490.
8. Daub, M. E., Herrero, S., and Chung, K. R. 2005. Photoactivated perylenequinone toxins in fungal pathogenesis of plants. FEMS Microbiol. Lett. 252:197-206.
9. Dekkers, K. L., You, B. J., Gowda, V. S., Liao, H. L., Lee, M. H., Bau, J. J., Ueng, P. P., and Chung, K. R. The Cercospora nicotianae gene encoding dual O-methyltransferase and FAD-dependent monooxygenase domains mediates cercosporin toxin biosynthesis. Fungal Genet. Biol. (In press).
10. Denslow, S. A., Walls, A. A., and Daub, M. E. 2005. Regulation of biosynthetic genes and antioxidant properties of vitamin B6 vitamers during plant defense responses. Physiol. Molec. Plant Pathol. 66:244-255.
11. Diwu, Z. 1995. Novel theraputic and diagnostic applications of hypocrellins and hypericins. Photochem. Photobiol. 61:529-539.
12. Ehrenshaft, M., Bilski, P., Li, M., Chignell, C. F., and Daub, M. E. 1999. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc. Natl. Acad. Sci. USA 96:9374-9378.
13. Ehrenshaft, M., and Upchurch, R. G. 1991. Isolation of light-enhanced cDNA clones of Cercospora kikuchii. Appl. Environ. Microbiol. 57:2671-2676.
14. Hayashi, K., Schoonbeek, H. J., and De Waard, M. A. 2002. Bcmfs1, a novel major facilitator superfamily transporter from Botrytis cinerea, provides tolerance towards the natural toxic compounds camptothecin and cercosporin and towards fungicides. Appl. Environ. Microbiol. 68:4996-5004.
15. Herrero, S., and Daub, M. E. 2007. Genetic manipulation of vitamin B-6 biosynthesis in tobacco and fungi uncovers limitations to up-regulation of the pathway. Plant Sci. 172:609-620.
16. Okubo, A., Yamazaki, S., and Fuwa, K. 1975. Biosynthesis of cercosporin. Agr. Biol. Chem. 39:1173-1175.
17. Shim, W. B., and Dunkle, L. D. 2003. CZK3, a MAP kinase kinase kinase homolog in Cercospora zeae-maydis, regulates cercosporin biosynthesis, fungal development, and pathogenesis. Mol. Plant-Microbe Interact. 16:760-768.
18. Spikes, J. D. 1989. Photosensitization. Pages 79-110 in: The Science of Photobiology. K. C. Smith, ed. Plenum Press, NY.
19. Taylor, T. V., Mitchell, T. K., and Daub, M. E. 2006. An oxidoreductase is involved in cercosporin degradation by the bacterium Xanthomonas campestris pv. zinniae. Appl. Environ. Microbiol. 72:6070-6078.
20. Upchurch, R. G., Rose, M. S., Eweida, M., and Callahan, T. M. 2002. Transgenic assessment of CFP-mediated cercosporin export and resistance in a cercosporin-sensitive fungus. Curr. Genet. 41:25-30.
21. Upchurch, R. G., Rose, M. S., Eweida, M., and Zuo, W. 2005. Expression of the cercosporin transporter, CFP, in tobacco reduces frog-eye lesion size. Biotechnol. Lett. 27:1543-1550.
22. Valenzeno, D. P., and Pooler, J. P. 1987. Photodynamic action. BioScience 37:270-275.
23. Ververidis, P., Davrazou, F., Diallinas, G., Georgakopoulos, D., Kanellis, A. K., and Panopoulos, N. 2001. A novel putative reductase (Cpd1p) and the multidrug exporter Snq2p are involved in resistance to cercosporin and other singlet oxygen-generating photosensitizers in Saccharomyces cerevisiae. Curr. Genet. 39:127-136.
