Introduction
Among the most notable emergent arthropod vector-plant virus complexes is the whitefly Bemisia tabaci (Genn.) and genus Begomovirus (Family, Geminiviridae) for which B. tabaci is the only insect vector. Begomoviruses have a circular, single-stranded DNA (ssDNA) genome. They are co-adapted with B. tabaci by which they are transmitted in a circulative, persistent manner. Begomovirus-vector complexes exhibit absolute transmission specificity within the viral genus/whitefly species, however, there is some evidence that transmission competency, or transmission efficiency, can vary. In general greater competency has been observed for begomovirus-B. tabaci complexes that share a common phylogeographical origin.
The increased importance of begomoviruses as new and emerging plant viral pathogens is directly related to the adaptive capacity of B. tabaci and its ability to exploit agricultural systems, which increasingly impose upon their subtropical habitats. B. tabaci has long confounded systematists, undermined agricultural production, and proven difficult to control partly because of its propensity to develop insecticide resistance.
This brief feature article will center on the recent history of the B. tabaci complex and the increasing importance of B. tabaci as a pest and virus vector to agriculture. It also will explore the development of the ‘biotype concept,’ including genetic and phenotypic variability, and the significance of these factors to pest/vector outbreaks and epidemics caused by whitefly-transmitted geminiviruses.
Whiteflies
Whiteflies are classified in the family Aleyrodidae (Sternorrhyncha: Hemiptera/s.o. Homoptera). The closest relatives to whiteflies are aphids, mealybugs, psyllids, and scales, all which feed using piercing and sucking mouthparts, which are specialized for feeding in plant phloem. Whiteflies are unique among their close relatives in that they utilize haplo-diploid reproduction, in which fertilized eggs give rise to females, and males are produced from unfertilized eggs. Thus all male offspring inherit only the maternal genome, whereas female offspring inherit genetic material from both parents. The male: female sex ratio in whitefly populations is regulated through an increased production of females when males are highly abundant. Conversely more male offspring are produced when males are in short supply, owing to the greater number of unfertilized eggs, thereby increasing the number of males, which in turn results in more female offspring being produced.
Whiteflies and other homopterans that feed exclusively on phloem sap are well known for harboring their own unique eubacterial endosymbionts, which are referred to as primary symbionts. Each whitefly species and the primary symbiont have been shown to exhibit a congruent evolutionary relationship, owing to their ancient association and now obligate, mutualistic relationship. Primary symbionts are associated with the reproductive organs of females (ovaries) and are inherited by the subsequent generation of offspring through the egg. The main function of the primary symbiont is thought to be the synthesis of amino acids, whose genes are encoded on bacterial plasmids. By analogy with aphid endosymbiont function, plasmids code for the production of essential amino acids that are naturally in short supply in phloem sap. In turn the whitefly host provides shelter and metabolic resources that have enabled (or resulted in) the reduction in size and complexity of the primary symbiont genome. Such obligately mutualistic relationships are believed to have initiated millions of years ago following a single bacterial infection event, and made it possible for homopterans to specialize in plant vasculature feeding. As such, primary endosymbionts are host-specific (based on the 16S ribosomal RNA gene) at the level of species, and this has generally been found to be the case for B. tabaci haplotypes. Other types (genera/species) of bacteria also may be associated with homopterans and although they have not been as widely studied, they are thought to contribute to environmental adaptation and ecological fitness. Still, other bacteria have been associated with B. tabaci, which are ‘parasitic,’ causing reproductive aberrations in B. tabaci.
| |

Fig. 1. Bemisia tabaci (Gennadius) whitefly adult. |
|
The Emergence of the Whitefly Bemisia tabaci (Gennadius) Pest and Vector
Among whiteflies, the B. tabaci complex is the most widely distributed and the most economically important worldwide. Both B. tabaci adults and nymphs can cause feeding damage, and the adults transmit several genera of plant viruses that undermine cultivated food and fiber production.
From the mid-1930s to the 1960s frequent outbreaks of B. tabaci were widely reported worldwide in vegetable and fiber crops grown in the tropics and subtropics. Infestations and/or virus-like diseases occurred in cotton in Sudan, affected vegetable crops in India and cassava in Africa, caused yield losses in soybean and tomato in Brazil, and leaf crumpling symptoms in cotton crops in Arizona and California, USA. The scientific literature reveals that much attention to these outbreaks was centered on pesticide solutions to reduce crop damage. In addition, the growing importance of B. tabaci as a new pest stimulated life history studies, cataloging of natural enemies, and epidemiological studies to address the increasing role as a vector of plant viruses. During the past thirty or so years, the importance of B. tabaci as a vector of plant viruses has continued to increase dramatically, to the extent that this once obscure whitefly is now among the most well known invasive insect and vector of plant viruses.
During 1975-1990 begomoviruses notably emerged widely as damaging viral pathogens in crop species throughout the tropical world and in mild temperate and Mediterranean locales. Several different genera of whitefly-transmitted viruses have been accidentally introduced to non-native locations, where they have rapidly established often causing more damage than the endemic viruses. The introduction of an exotic, pesticide resistant biotype (B) of B. tabaci to many world locations during 1986-1992 drew increasing attention to the pest and vector status of B. tabaci.
Clearly, B. tabaci and the plant viral pathogens it transmits are no longer restricted to their native habitats or contained by natural geographic boundaries. Today, B. tabaci is considered among the most invasive and economically damaging insects to agriculture, spanning food and fiber crops, and certain nursery grown ornamentals, with the ability to colonize more than 500 plant species. Increased monoculture cropping, reduced genetic variation in cultivated species, widespread insecticide use in agriculture, and the international transport of live plants are among the most substantial factors that have contributed to the growing worldwide importance of indigenous and exotic B. tabaci and the plant viruses they transmit.
Whitefly-transmitted Geminiviruses
For perspective, it is important to recognize that the Geminiviridae was established in only 1978. Further, members of this unusual plant virus family contain a circular, single-stranded DNA genome, and so they are not like the majority of plant viruses, which contain RNA. Geminiviruses genera (now 4) are grouped by arthropod vector (leafhopper, planthopper, treehopper, or whitefly) and whether they infect dicotyledonous or monocotyledonous plants. When the Geminiviridae was established, there were fewer than ten recognized species. Indeed, many other virus-like diseases were suspected to have begomoviral etiology but proof had not yet been provided. Even so, it was surmised that the whitefly-transmitted geminiviruses (subgroup III) (now the genus Begomovirus), likely comprised quite a large number of distinct species, owing to the apparent wide distribution of the so-called ‘rugaceous’ or ‘yellowing’ diseases associated with whitefly infestations. The hallmark foliar symptoms of these whitefly-associated virus-like pathogens were characteristic curling, mosaic, and bright yellow or yellow-green discoloration, which set them apart from their more traditionally studied, RNA virus counterparts. Disease symptoms were often most striking in native or ‘endemic’ dicotyledonous plants (eudicots) from which they presumably originated, compared to those of cultivated plant species, which were nonetheless often dramatic.
Although the first electron micrographs were not produced until the early 1970s, several groups in the tropical Americas and United Kingdom simultaneously revealed the earliest evidence for an unusual ‘geminate’ particle morphology, from which they were hypothesized to constitute a new and unusual type of plant virus. The discovery that they contained circular, single-stranded DNA (ssDNA) genomes fueled widespread interest, and the timely development of recombinant DNA technologies permitted the cloning and sequencing of several begomoviral genomes within a decade after their discovery. Even so, by 1980 only a handful of whitefly-transmitted geminiviruses (genus, Begomovirus) had been sequenced. However, this was sufficient to facilitate the reconstruction of phylogenetic relationships and hence taxonomic assignment, and the development of primers when polymerase chain reaction (PCR) technology was developed in the 1980s. However, only in the late 1980s-early 1990s, did it become apparent that the whitefly vector was widely variable and phenotypically plastic such that in the face of rapidly changing practices in crop production, such extreme adaptability would directly influence the diversification and emergence of a plethora of new begomoviral species in cropping systems, worldwide. Unimaginable was the possibility that by the year 2006, the genome sequence would be determined for several hundred begomoviral species (International Committee on the Taxonomy of Viruses database).
The emergence and in some instances introduction of certain begomoviral species into new habitats is directly correlated to high population levels of their whitefly vector. Viruses once endemic to native plant species are rapidly making their way into cultivated monoculture settings, in which they rapidly diversify and become highly fit pathogens in crop species. Begomoviruses have only recently been shown to readily and rapidly diversify by employing inter-specific recombination, which can occur when two or more viruses infect nuclei (replicate) of the same plant. And bipartite begomoviruses also are capable of ‘reassortment,’ or swapping their DNA-A and DNA-B components, provided that the iterated (repeated) sequences, to which the virus-specific replication-associated protein (REP) binds to initiate virus replication, are sufficiently compatible. This is sometimes seen among closely related begomoviral species, but sometimes even more distantly related virus components are compatible when one or the other has undergone recombination in this region, even though otherwise they may be distantly related.
 |
|
 |
|
Fig. 2. ‘Delgado’ bean inoculated with Squash leaf curl virus (Arizona isolate 1990). |
|
Fig. 3. Winter squash infected with Squash leaf curl virus (Arizona 2001). |
 |
|
 |
|
Fig. 4. Begomovirus symptoms in tomato shortly after the introduction of the B biotype (Pernambuco, Brazil 2001). |
|
Fig. 5. ‘Pinto’ bean plants inoculated with infectious clones of Bean calico mosaic virus originating from Sonora, Mexico (Arizona 1998). |
 |
|
 |
|
Fig. 6. Symptoms in cotton of a close relative to Cotton leaf crumple virus (Guatemala 1992). |
|
Fig. 7. Symptoms of Cabbage leaf curl virus (native to Florida) in cabbage seedlings. |
 |
|
 |
|
Fig. 8. Tomato yellow leaf curl virus in tomato seedlings (Sinaloa, Mexico 2005). |
|
Fig. 9. Pepper infected with Pepper golden mosaic virus (Sonora, Mexico 1989 isolate). |
Clearly, as whitefly vector population levels continue to remain robust and persistently associated with cropping systems, particularly in subtropical and mild climate locales, new begomoviral species will emerge and cause damaging diseases in food and fiber crops. Indeed, in certain locations new viral species may often be discovered every several years, with the viruses that were previously important disappearing nearly as quickly as they emerged. This pattern has been particularly true in parts of Brazil, Guatemala, and Mexico where vegetables are grown for export, typically for winter markets in the US and Europe. Undoubtedly, the development of virus-resistant crop varieties should be a high priority, employing viable, effective breeding and/or transgenic approaches. Further, novel strategies that rely on the interruption of whitefly cues involved in host finding, and interference with conserved interactions in the virus-vector transmission pathway are among other feasible avenues that should be rigorously explored to collectively control virus diseases and reduce pesticide use to control whiteflies. Anything less is likely to lead to even greater difficulties to producers, and an increased requirement for expensive inputs to enable vegetable, fruit, fiber, and ornamental crop production in sufficient quantities to sustain human needs.
Vector Transmission Specificity and Competency
Whitefly-virus vector specificity is a highly conserved feature of begomoviruses. Worldwide they are transmitted by the single species B. tabaci. This can be corroborated biochemically by the high degree of conservation maintained in the begomoviral capsid protein, the only viral-encoded protein required for whitefly-mediated transmission. That there is no robust evidence that a whitefly species other than B. tabaci can serve as a begomovirus vector further supports the proposed ‘single species’ nature of B. tabaci. Indeed, the genus begomovirus could well be seen as the arbiter to the lumping or splitting of the species. Further, the B. tabaci primary endosymbiont that synthesizes the 60S heat shock protein, apparently essential for virus transmission, also is co-evolved with its whitefly host.
| |
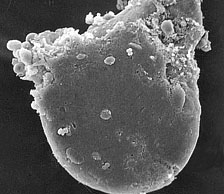
Fig. 10. Bacteriosome consisting of an aggregate of 60 to 90 cells, called bacteriocytes, that contain the primary symbiont of B. tabaci. |
|
Even though transmission specificity seems simplified to involve ‘one virus-one vector,’ it is important to emphasize that subtle selection forces influence and modulate the transmission processes at the coevolving virus-vector interface. Such selection is envisaged to be mediated by host preference combined with vector competency, neither of which is well studied mechanistically. Vector host preferences and transmission competency may become irrelevant under high vector pressures, the latter of which is typified by cropping systems converging with endemic plant-virus complexes. The emergence of new biotypes and/or new begomoviral species originating from native hosts appears to occur as these complexes are ‘test driven’ when contact is made with cropping systems, where vector population levels and the frequency of viruliferous individuals are extremely high, facilitating the adaptation of the virus to new hosts with surprising rapidity.
 |
|
 |
|
Fig. 11. Macroptilium lathyroides naturally-infected with a native begomovirus species (Guatemala 2001). |
|
Fig. 12. Begomovirus symptoms in Euphorbia species (Brazil 2001). |
The evolutionarily conserved virus-vector-endosymbiont relationship, together with an unexpected ability to adapt to environments altered in the interest of agriculture sets apart begomovirus-whitefly vector complexes from most other plant viruses and their arthropod vectors. In addition, a co-evolved ‘transmission pathway,’ or route by which virions circulate in the vector, is conserved across all B. tabaci variants and begomoviruses, effectively permitting whitefly-mediated transmission of any begomovirus by any B. tabaci, providing it can feed on the particular virus host. This is providing an opportunity to dissect the cellular and molecular basis of transmission specificity and for the apparently subtle differences in vector competency, which has been alluded to by several studies. The conserved specificity has its basis in the viral coat protein, (the most conserved viral protein for the genus), putative receptors in the gut and salivary glands of the whitefly, a primary endosymbiont-encoded heat shock protein (HSP 60), and probably several whitefly-encoded factors. Precisely how these intricately interactive, multi-kingdom components contribute to the circulative passage of virions, unimpeded, across the otherwise selective gut and salivary gland membrane barriers of this whitefly species remains an unresolved question.
| |
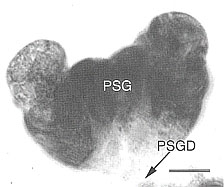
Fig. 13. Primary salivary gland of B. tabaci. |
|
The Bemisia tabaci Complex: Biotic and Genetic Conundrums
Phenotypic variability and plasticity. The B. tabaci complex is an excellent example of a cryptic species for which genetic variation can be demonstrated in the absence of morphological variation in adults or pupae, the latter which is used by taxonomists to identify whiteflies. Surprisingly, biotic (phenotypic behaviors) and genetic variants exhibit no corresponding morphological characters that permit their discrimination.
| |
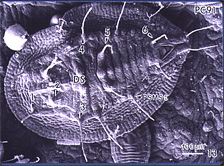
Fig. 14. Pupal case for Bemisia tabaci used for identification to species. Few if any differences are seen for different biotypes or haplotypes. |
|
A further complication involving pupal morphology is that B. tabaci responds to plant surface topology by alteration of the sizes and shapes of setae, hairs, pores, and waxy protrusions on the pupae, making it visibly adaptive, and at times difficult to correctly identify it to species. These collective characteristics have earned B. tabaci the reputation of a taxonomically confounded insect with behavioral and genetic features that do not overtly correlate with external anatomical adaptations. Instead, adaptation appears to be regulated at the biochemical level, facilitating plasticity in responsiveness to environmental associated variables.
The natural plant hosts of B. tabaci are represented by a wide array of annual or perennial dicotyledonous species (now, Eudicots) often found in the dry-subtropical forest understory, and along riverbanks and washes in desert environments, throughout the subtropics, tropics, and mild or Mediterranean climates where prolonged freezing temperatures are rare or nonexistent. This whitefly species colonizes over 500 plant species, including endemic and cultivated species, and is referred to as polyphagous. However, some B. tabaci populations are moderately polyphagous, or host-restricted, or monophagous (typically colonizing one to three related genera or species). Typically the polyphagous types of B. tabaci are most frequently associated with disease and pest outbreaks. However, in Africa and India some B. tabaci are host-adapted to cassava, indicating that monophagous populations also can be problematic in food production. B. tabaci is important in these areas because it is the vector of begomoviruses that infect cassava, and less so because of colonization pressure. Cassava mosaic disease is caused by a number of begomovirus species that are indigenous to Africa, whereas in India a distinct (and divergent) species infects cassava, also causing mosaic symptoms.
It was Dr. Julio Bird who first recognized phenotypic variation in the B. tabaci found on the island of Puerto Rico. Bird recognized a polyphagous variant, the ‘Sida race,’ and a monophagous variant, ‘the Jatropha race,’ based on natural and experimental host preferences observed for each. In 1975, L. Russell (USA) and A.S. Costa (Brazil) took notice of Bird’s discovery, pointing out that B. tabaci in Brazil was never observed colonizing cassava, a plant species native to South America, whereas, in Africa B. tabaci widely colonized cassava, following its introduction by European colonists as a staple.
Although the genetic basis for the extraordinary adaptability of B. tabaci is not known, the species has become widely recognized for the damage it causes in subtropical and tropical cropping systems. B. tabaci effects crop losses through direct feeding damage, honeydew contamination of plant parts, and the transmission of plant viruses. Indirectly, downstream problems include exacerbating or facilitating diversification of begomoviruses, which has the potential to lead to host-shifts, increased virus virulence, and the emergence of new viral species, both in cultivated and endemic species.
Other behavioral phenotypes that are recognized among B. tabaci biotypes (and less well characterized variants that are not yet considered biological types) include adaptations for variation in fecundity (extremely high = hundreds per plant, to low = 2-6 per plant), dispersal behavior (long flights or short distance ‘hopping’), rapid or slow responsiveness to disturbance, induction of phytotoxic responses in certain hosts (e.g., silvering of Cucurbita species, irregular ripening of tomato, white stalk of broccoli), endosymbiont complement, and differential sensitivity to insecticides.
Genetic polymorphisms. General esterases were the first B. tabaci proteins explored in the search for genetic (biochemical) polymorphisms, providing corroborative support for observed phenotypic variation. The general esterase approach, combined with detailed biological and morphological studies, was used to characterize in the same laboratory setting B. tabaci representing different geographic locations, worldwide. Later these and additional populations were characterized using random amplified polymorphic DNAs (RAPDs) and directly by DNA sequencing of molecular markers.
| |
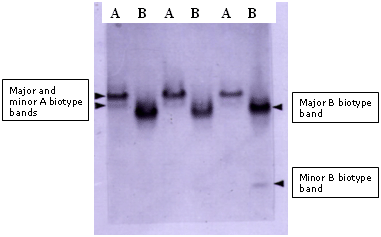
Fig. 15. General esterase banding patterns for the A and B biotypes that allowed for their initial. |
|
The study by Costa and Brown in 1991 established the A and B biotype nomenclature. The initially arbitrary alphabetical designations were used to refer to the ‘local’ B. tabaci population from cotton in Arizona, U.S., and a population found unexpectedly colonizing poinsettia, which produced different esterase patterns. These protein polymorphisms were subsequently supported by differences in host range and fecundity, and evidence that the poinsettia type induced phytotoxic symptoms in pumpkin, whereas, the Arizona cotton colony did not. The distinguishing esterase patterns were applied to differentiate the biologically distinct populations, and thereafter, they were referred to as the A (local cotton colony) and B (poinsettia colony) biotypes.
| |
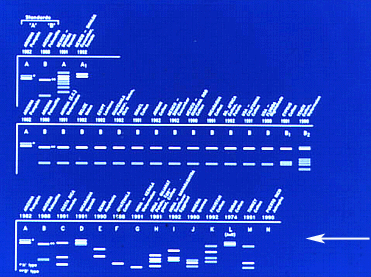
Fig. 16. First evidence for widespread esterase polymorphisms for the B. tabaci complex. |
|
With the analysis of more populations collected in the American Tropics, additional novel esterase patterns were observed, suggesting that this whitefly was more polymorphic than had been realized. With the discovery of each new esterase pattern, the respective population was assigned a sequential letter of the alphabet, which unfortunately, led to the misnomer that each represented a distinct biotype.
| |

Fig. 17. Symptoms of squash silverleaf in pumpkin (left) and whitefly free, unaffected pumpkin plants (right). |
|
While not all collections could be established in laboratory colonies, those that could be established were the subject of a comparative study to characterize them with respect to partial host range, fecundity, morphological variability of pupae, size of adults, potential for reciprocal crossing, ability to induce silvering in pumpkin, and virus transmission. The results revealed a surprising array of behaviors, including mating incompatibility, differential vector competency, differences in fecundity and ability to induce the silvering phenotype, and size differences for adults. As well, insecticide resistance was shown to be variable with respect to types of chemistries when examined for selected populations. However, it was not possible to identify informative morphological characters using morphometric or classical taxonomic approaches with pupae.
Genetic variability was next explored using RAPDs-PCR and isozyme analyses with results corroborating the extensive intraspecific variation first revealed through protein polymorphisms. The greatest drawback to RAPDs for resolving population structure was the lack of well-studied reference populations, which greatly hindered the interpretation of the results. Another problem was the lack of reproducibility of patterns between laboratories, and so even though a fair number of populations were examined this way, the data sets did not add significantly to the knowledge base. Even so, based on results of RAPDs and isozyme analysis and the inability to demonstrate gene flow in reciprocal crosses of the A and B biotypes, one laboratory concluded that the B biotype should be considered a separate species of Bemisia and proposed the name B. argentifolii. The specific epithet has not, however, become generally accepted, nor has it been approved by the nomenclature committee of the Entomological Society of America.
Molecular markers and phylogeographic distribution. Genetics studies next examined DNA variability within the B. tabaci complex directly, or by sequencing selected genes. The first study of this type explored first the mitochondria (mt) 16S ribosomal RNA gene (rDNA), followed by the mt cytochrome oxidase I (mtCOI) sequence. For both markers, phylogenetic analyses revealed a robust phylogeographic grouping of haplotypes into one of seven major monophyletic clades that are nearly equally divergent (inter-clade divergence) from one another at ~10%. All exemplars grouped with a strong phylogeographical basis, with one exception, the B biotype. This is because by then, this invasive biotype had become widely distributed via international trade of ornamental plants. Of the two markers, the mtCOI sequence has proved to be the more robust, revealing the greatest nucleotide divergence between and within populations, and consequently making it a highly useful molecular marker.
Phylogenetic analysis of representative B. tabaci mtCOI sequences for an extensive worldwide collection has revealed that B. tabaci haplotypes group into one of ~eight (or nine) predominant clades, or ‘phylogeographic groups’. Based on these results, B. tabaci haplotypes cluster together based on their native geographical origin and natural range. This holds true for all B. tabaci examined to date except for two exotic haplotypes (B and Q), which are known to have been introduced and become established in non-native locations recently. The application of the mtCOI sequence as an ‘informative’ molecular marker has made possible the determination of the origins for the B and Q exotic (or introduced) biotypes, because they are ‘out of place,’ or highly divergent from the native haplotypes that inhabit the zones which they have invaded.
Each mtCOI sequence groups in a single ‘major mtCOI clade’ with a geographical basis in extant origin. The major clades diverge approximately equally from each other, making it impossible to root the mtCOI tree, and suggesting that the respective haplotypes have been geographically and/or reproductively isolated from one another for some time. Interestingly, however, ‘within clade divergence’ varies, depending on the particular clade. For example, within the major American Tropics clade, the North America/Caribbean and South America sister clades exhibit the least amount of divergence at ~5-8% (collectively), whereas B. tabaci haplotypes from throughout continental African are highly divergent (~16-26%), with the Asian haplotypes exhibiting the second greatest divergence at ~12-20%. These results suggest that the B. tabaci complex m4y have first diversified in Africa, from where certain populations spread to Asia and vicinity. That extant American Tropics haplotypes are the least genetically divergent (extant populations) worldwide, suggests minimal radiation into the western hemisphere, or possibly abundant ancestral radiation, followed by the survival of only a few haplotypes.
Importantly, mtCOI analysis indicates that the B and Q biotypes are native to the vicinity of the Middle East-eastern Africa, and the Mediterranean region, including North Africa, respectively. This is based on their placement in the major Mediterranean/North African/Middle Eastern clade, albeit, they are each members of different sister clades. The mtCOI analysis indicated that the B biotype did not at all stand out as a separate taxon, when considered in the backdrop of a worldwide collection of B. tabaci haplotypes. Similar relationships have been elucidated when large sample sizes of globally representative collections have been subjected to isozyme or RAPDs analyses. These comprehensive analyses also revealed close to borderline genetic distances, with respect to the predicted values for species cutoff, further corroborating the diversity and complex genetics of B. tabaci, and underscoring the non-uniqueness of the B biotype in global context of diverse exemplars of this species. Further, when molecular information regarding the predicted origin of the B biotype was made available following its invasion in the Western Hemisphere during 1990-94, this knowledge greatly aided those prospecting for biological control agents (parasitoids, predators) by pinpointing the locales from which co-evolved natural enemies might best be collected for laboratory assessment, thereby avoiding the necessity of a worldwide exploration effort.
| |
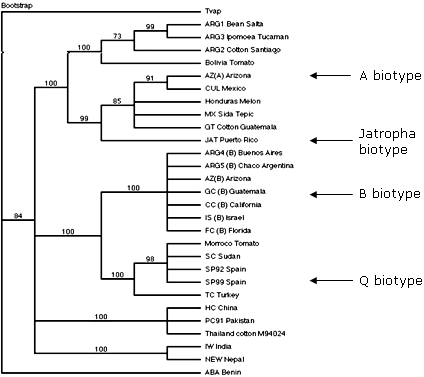
Fig. 18. Phylogeographic distribution of four major biotypes of the B. tabaci complex based on the mitochondria cytochrome oxidase I sequence. |
|
The Growing Importance Of Highly Adaptable And Invasive Biotypes
The increased importance of the B. tabaci complex and associated virus disease outbreaks in vegetables and fiber crops is largely due to recent changes in the way crops are now produced. Less than one hundred years ago, these crops were cultivated in relatively small acreages in semi-temperate or Mediterranean climates, and markets and processing facilities were nearby. In contrast, large-scale food production is practiced both in mild-temperate climates under irrigation, and in subtropical parts of the world where 2 or more crops per year can be produced. In both types of cropping, fields are left fallow only during the short winter or rainy season, respectively. Further, contemporary agriculture increasingly employs a genetically narrow germplasm base in which abiotic and biotic stress resistance has been selected against during the breeding in search of high yielding varieties and/or cultivars with the most desirable horticultural traits. High inputs, including fertilizers and pesticides, are required for this high-intensity food production, leading to non-target exposures and subsequent development of insecticide resistance. Finally, international trade has increased substantially for live plants and plant parts, providing conduits for whiteflies and/or virus to disperse or be carried freely between once isolated regions and continents. Among the unfortunate outcomes has been the selection of ‘biological types’ of B. tabaci that are adapted to modified habitats, which provide ample resources to support reproduction and sustain large population sizes over time. The B. tabaci complex is a colonizer of ‘marginal habitats,’ producing few offspring when the supply of food supply is meager, but having the capacity to develop large population sizes when resources become available, typically at the cessation of the rainy season in subtropical or Mediterranean climates when plant growth is flush and widely available for reproduction.
Presently, as indicated, two main exotic biotypes of B. tabaci, termed the B and Q types, have become widespread in ornamental plants and agricultural cropping systems, nearly worldwide. The B biotype is by far the most widely distributed, but the recent introduction (2004-present) of the Q biotype from the Mediterranean region into North (Mexico, U.S.) and Central America (Guatemala), and multiple locations in Asia (China, Japan, among others), points to the need for diligent tracking and self-monitoring on imported plants, and the legalized regulation of the transport of this insect, if need be. If neither precaution is implemented rigorously for the long term, the unrestricted movement of plants infested with B. tabaci is expected to lead to the introduction of additional highly damaging, exotic biotypes. Further the potential for the concurrent introduction of exotic plant viruses carried by invasive, viruliferous B. tabaci poses an equally serious threat to vegetable and fiber production in the western hemisphere, including the southern and southwestern U.S. and Mexico. To date, Tomato yellow leaf curl virus (Geminiviridae) and Cucurbit yellow stunting disorder virus (Closteroviridae), both hailing from the Middle East/Mediterranean region now have been introduced into the U.S. and Mexico, with the precise mode of transport as yet undetermined.
| |

Fig. 19. First symptoms of Cucurbit yellow stunting disorder virus in melon crops in Yuma, Arizona during the fall 2007. |
|
The B and Q Biotypes
The unwitting introduction of the B biotype in the U.S. and Caribbean in 1986-1990 ferried with it the first widespread awareness of B. tabaci. The rapid establishment of this exotic biotype was marked by outbreaks of phytotoxic symptoms. Phytotoxic-like disorders were observed in cucurbits, cole crops, and tomato plantings in the western US, Mexico, and the Caribbean region. Eventually these symptoms became diagnostic for the presence of the now well-studied B biotype of B. tabaci. Coincident with the rapid invasion of the B biotype in the Americas was the emergence of uncharacterized plant viruses, later identified as members of the genus, Begomovirus. Particularly notable were diseases of cabbage, cucurbits, pepper, and tomato, which are not preferred hosts of the native B. tabaci. During 1991, B biotype populations exploded, reaching unprecedented levels in irrigated cropping systems of the southwestern U.S., and uncontrollable infestations throughout the southern U.S. Soon, the B biotype spread to the Caribbean region and American Tropics, reaching South America by 1994. Australia, China, Egypt, Mediterranean region of Europe, Israel, Japan, Pakistan, and Turkey reported B biotype outbreaks during 1990-1994, making it the first B. tabaci biotype to become recognized as a cosmopolitan pest and vector. As rapidly, its propensity to develop insecticide resistance became apparent, and so efforts were undertaken to establish monitoring and resistance management programs.
The Q biotype, characterized first from Spain, and its close relatives (Europe/North Africa/Middle East clade) are indigenous to the Mediterranean region. Vegetable crops, including tomato, are preferred hosts. Members of this clade are moderately-to-widely polyphagous, and are (or were) extant in Europe, northern and western Africa, Turkey, Morocco, Sudan, Egypt, Israel, and are found as far south as Uganda. The Spanish Q was the predominant haplotype in Spain prior to the B biotype invasion. By the early 1990s the B biotype was widespread in the habitat of the Spanish-Q and Q-like relatives in the region, including in Turkey where it displaced the local ‘TC’ variant. However, as discussed below, the Q biotype is now displacing the B biotype in some of these same areas.
The Q biotype is presently the major B. tabaci pest and vector in Mediterranean climates where it originated, and also is problematic in commercial greenhouses producing ornamental plants. The partial displacement of the B biotype by a Q-like relative in Israel is also thought to be due to differential insecticide resistance in the two populations, given that subsequent outbreaks of this close Q relative coincided with neonicotinoid insecticide use to control the B biotype. Because the Q biotype also has a broad host range that includes cultivated and uncultivated species, and has exhibited resistance to the insecticides that control the B biotype in Spain and Israel, it poses a renewed threat. Thus, clearly the B and Q biotypes display differential capabilities for detoxifying insecticidal chemistries commonly used for whitefly control. It is thought that the management of B biotype using certain types of compounds has led to the development of resistance in the Q biotype to the latter chemistries, even though the B biotype remains held in check. Consequently, insecticide resistance is an important determinant of fitness in presently problematic B. tabaci and will likely be for other biotypes that upsurge in the future.
Protected vegetable production in controlled environment facilities is a rapidly growing industry in Spain and in the U.S., Mexico, and Central America, and whiteflies and plant viruses they vector are an emerging problem in the industry. If the Q biotype becomes established in greenhouses, it could spread to cotton and vegetable crops in nearby fields. If this occurs, it is likely that producers will be unable to control the Q biotype using insecticides that are presently effective against the B biotype, and which are making food and fiber production possible. Only in 2005 did the Q biotype became prevalent on ornamentals, and from this commodity, this invasive biotype was subsequently exported to China, Japan, Mexico, and at least 22 states in the U.S., with the first U.S. report being in Arizona on poinsettia plants. Had molecular diagnostics tools been implemented at the ports of entry, neither the B nor Q biotypes would have become so quickly widespread, and the introduction of the Q biotype likely could have been avoided altogether.
The Q biotype continues to be imported from offshore ornamentals nurseries, and federal regulations do not prohibit the importation. This could have further significance to U.S.-exported plant products if other countries decide to regulate this insect. Biotypes of B. tabaci are not regulated in the U.S. because, as noted above, many prevalent whitefly genera, including B. tabaci are difficult to distinguish using classical taxonomic approaches that rely on differential morphological traits. Discerning between whitefly species that are common in nursery and floral crops, including B. tabaci biotypes, requires implementation of molecular diagnostics. Although molecular tools have been widely implemented to track the dispersal of the B and Q biotypes, and to study the diversity of B. tabaci worldwide, these approaches have not yet been implemented at U.S. Ports of Entry.
Even with molecular markers, however, it has not been possible to predict the precise origin, or to reconstruct the evolutionary history for the B. tabaci complex, which has radiated worldwide in the tropics. However, the recent development of molecular markers and application of population genetics approaches have made possible a glimpse of the intriguing biogeographical relationships shared among this fascinating and troublesome insect species. Additional studies are underway that will increase our understanding of the natural history, the underlying virus-vector specificity, and the population genetics and evolutionary biology of this economically important and scientifically interesting species complex.
Conclusions
Well-known biotypes of B. tabaci are readily distinguished using molecular diagnostics. The most definitive and informative approach utilizes PCR amplification and DNA sequencing of a fragment of the mtCOI, yielding useful phylogeographical information regarding the extant origin of particular haplotypes (and biotypes). Subsequent mtCOI sequence comparisons using extensive reference to public and unpublished databases, permits discrimination not only between biotypes of B. tabaci, but also makes possible identification of other whitefly species that colonize the same hosts and may inadvertently be collected with B. tabaci. Even so, the ability to distinguish common biological types has not prompted the establishment of quarantines against the most tenacious B. tabaci variants, because morphologically based identification remains the most practical means for general identification of insects at ports of entry. Indeed, a major shortcoming to advancing the biotype concept is the shortage of biological data available for the growing number of well-defined genetic variants. There is a need for quarantine-level insectaries in which whitefly colonies can be maintained in isolation and used to carry out rigorous life history, host range, mating compatibility, and virus-vector studies, among others. Soon genomics-based technologies will make possible the linking of phenotypic and genetic variability to gene expression patterns. Identifying regions of cross-kingdom proteome conservation will reveal orthologous arthropod genes with functionalities in adaptive life history traits, reproductive isolation mechanisms, competitiveness/invasiveness versus benign colonization, and cellular and molecular co-adaptation that confer virus-vector specificity and influence transmission competency, all presently unidentified. Such approaches are expected to enable predictions of volatile or benign biotype invasions, encourage development of more directed, environmentally sound ‘pest’ and ‘vector’ management strategies, and fuel scientific inquiry that seeks to unravel to the next level the underpinnings of this minute phloem-feeder, an insect that for more than 120-140 million years has continued to perfect its ability to exploit inter-kingdom interactions with resounding success.
Selected References
Bedford, I. D., Markham, P. G., Brown J. K., and Rosell, R. C. 1994. Geminivirus transmission and biological characterization of whitefly (Bemisia tabaci) biotypes from different world regions. Ann. Appl. Biol. 125:311-325.
Berry, S., Rey, M. E. C., Rogan, D., Fondong, V. N., Fauquet, C. M., and Brown, J. K. 2004. Molecular evidence for distinct Bemisia tabaci geographically genotypes from cassava in Africa. Ann. Entom. Soc. Am. 97:852-859.
Bird, J., and Sanchez, J. 1971. Whitefly-transmitted viruses in Puerto Rico. Tech Paper, Agric. Exp. Station, Univ. of Puerto Rico 55:461-467.
Brown, J. K., Frohlich, D., and Rosell, R. 1995. The sweetpotato/silverleaf whiteflies: biotypes of Bemisia tabaci (Genn.), or a species complex? Ann. Rev. Entomol. 40:511-534.
Brown, J. K. 1994. The status of Bemisia tabaci (Genn.) as a pest and vector in world agroecosystems. FAO Plant Prot. Bull. 42:3-32.
Brown, J. K. 1990. An update on the whitefly-transmitted geminiviruses in the Americas and the Caribbean Basin. FAO Plant Prot. Bull. 39:5-23.
Brown, J. K. 2001. The molecular epidemiology of begomoviruses. Pages 279-316 in: Trends in Plant Virology. J. A. Khan and J. Dykstra, eds. The Haworth Press, Inc., NY.
Brown, J. K., and Czosnek, H. 2002. Whitefly transmitted viruses. Pages 65-100 in: Adv. Botan. Res. Academic Press, N.Y.
Brown, J. K., and Bird, J. 1992. Whitefly-transmitted geminiviruses in the Americas and the Caribbean Basin: past and present. Plant Dis. 76:220-225.
Brown, J. K., Coats, S., Bedford, I. D., Markham, P. G., Bird, J., and Frohlich, D. R. 1995. Characterization and distribution of esterase electromorphs in the whitefly, Bemisia tabaci (Genn.) (Homoptera: Aleyrodidae). Biochem. Genet. 33:205-214.
Brown, J. K., French, R., Rogan, D., and Bird, J. 2006. Biotic and mtCOI analysis for the Bemisia tabaci complex reveals extraordinary genetic variability, incomplete reproductive isolation, and a history of population expansions and invasions. (in preparation).
Brown, J. K., and Idris, A. M. 2005. Genetic differentiation of the whitefly Bemisia tabaci (Genn.) mitochondria COI and geographic congruence with the coat protein of the plant virus genus: Begomovirus. Ann. Entom. Soc. Am. 98:827-837.
Byrne, D. N., and Bellows, T. S. 1991. Whitefly biology. Ann. Rev. Entomol. 36:431-457.
Campbell, B. C., Steffen-Campbell, J. D., and Gill, R. 1994. Evolutionary origin of whiteflies (Hemiptera: Sternorrhyncha: Aleyrodidae) inferred from 18S rDNA sequences. Insect Mol. Biol. 3:73-89.
Cock, M. J. W. 1993. Bemisia tabaci, an update 1986-1992 on the cotton whitefly with an annotated bibliography. CAB IIBC, Silwood Park, UK, 78pp.
Costa, H. S., and Brown, J. K. 1991. Variation in biological characteristics and in esterase patterns among populations of Bemisia tabaci (Genn.) and the association of one population with silverleaf symptom development. Entomol. Exp. Appl. 61:211-219.
Costa, H.S., Brown, J. K., Sivasupramaniam, S., and Bird, J. 1993. Regional distribution, insecticide resistance, and reciprocal crosses between the `A' and `B' biotypes of Bemisia tabaci. Insect Sci. Applic. 14:127-138.
Czosnek, H., Ghanim, M., Morin, S., Rubinstein, G., Fridman, V., and Zeidan, M. 2001. Whiteflies: Vectors, and Victims (?) of Geminiviruses. Adv. Virus Res. 57:291-322.
Dennehy, T. J., DeGain, B. A., Harpold, V. S., Brown, J. K., Morin, S., Jeff A. Fabrick, J. A., Byrne, F. J., and Nichols, R. L. 2006. New whitefly biotype with high levels of resistance to insecticides. The University of Arizona - Cooperative Extension Report 32 pp.
Frohlich, D. R., Brown, J. K., Bedford, I., Markham, P. G. 1996. Mitochondrial 16S ribosomal subunit as a molecular marker in Bemisia, and implications for population variability. Pages 143-145 in: Bemisia 1995: Taxonomy, Biology, Damage Control, and Management. Intercept, Ltd., Andover, Hants, UK.
Gill, R. J. 1992. The morphology of whiteflies. Pages 13-46 In: Whiteflies: Their Bionomics, Pest Status, and Management. D. Gerling, ed. Intercept Ltd, Andover, Hants, UK.
Isakeit, T., Idris, A. M., Sunter, G., Black, M. C., and Brown, J. K. 2007. Tomato yellow leaf curl virus in tomato in Texas, originating from transplant facilities. Plant Dis. (accepted).
Legg, J. P. 1996. Host-associated strains within Ugandan populations of the whitefly Bemisia tabaci (Genn) (Hom, Aleyrodidae). J. Appl. Entomol. 120:523-527.
Martinez-Carillo, J. L., and Brown, J. K. 2007. First report of the Q biotype of Bemisia tabaci (Gennadius) in southern Sonora, Mexico. Phytoparasitica (accepted).
Qiu, B. -L., Coats, S. A., Ren, S. -X., Idris, A. M., Caixia, X. U., and Brown, J. K. 2007. Phylogenetic analysis (mtCOI) and evidence for generalized polyphagy for the Bemisia tabaci (Genn.) complex (s.o. Homoptera: Aleyrodidae) endemic to Southeast Asia reveals a high-diversity hot spot, second only to Africa. Progress in Natural Science (accepted for publication, August 2007).
Rybicki, E. P. 1994. A phylogenetic and evolutionary justification for three genera of Geminiviridae. Arch. Virol. 139:49-77.
Sseruwagi. P., Maruthi, M. N., Colvin, J., Rey, M. E. C., Brown, J. K., and Legg, J. P. 2006. Colonization of non-cassava plant species by cassava whiteflies (Bemisia tabaci) in Uganda. Ent. Exp. et Applic. 119:145-153.
