Robert S. Zeigler
Department of Plant Pathology, Kansas State University, Manhattan, KS 66506-5502. rzeigler@ksu.edu
Fernando J. Correa
Rice Project, Centro Internacional de Agricultura Tropical (CIAT), AA 6713, Cali, Colombia. f.correa@cgiar.org
Zeigler, R.S. and Correa, F.J. 2000. Applying Magnaporthe grisea population analyses for durable rice blast resistance. 2000. APSnet Features. Online. doi: 10.1094/APSnetFeature-2000-0700A
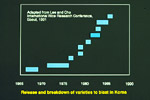
The blast disease of rice, caused by Pyricularia grisea Sacc., the anamorph of Magnaporthe grisea (Hebert) Barr, is the most widespread and damaging disease of cultivated rice, prevalent in both the tropics and temperate zones (22). The fungus may attack all above-ground parts of the rice plant, but typically leaf and panicle lesions are the most serious (Figures 1 and 2). For many years research focussed on the identification of pathogenic races and incorporating corresponding resistance genes into commercial varieties (17). However, durable resistance to the pathogen has eluded breeders and pathologists despite decades of concentrated breeding efforts and research. Typically, a variety released as blast-resistant shows signs of susceptibility after only very few seasons of cultivation in blast-prone environments (Figure 3).
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
| Click any image for enlargement. |
The search for the explanation of the rapid "breakdown" of blast resistance sparked one of the most contentious debates in modern plant pathology. The debate centered around the origin of the diversity of pathotypes, or races, detected in the field. At one extreme the pathogen was described as hypervariable, with the capacity to generate a seemingly endless array of new pathotypes from a single asexual spore (16). Thus, varieties evaluated for resistance to a single pathotype would be exposed to an infinite range of pathogenic variation once released into the field. A variety stood little chance of surviving under the onslaught of such variation, and the reasonable conclusion was that race-specific resistance to the pathogen could not yield durable resistance. This led to a major effort to develop race non-specific, or partial, resistance (1,13). At the other extreme, the pathogen was described as completely stable, with no new races generated even after years of culture in the laboratory (9,10). It is noteworthy that the proponents of hypervariability worked with isolates from Asia (the center of origin of rice), recently recovered from the field, while proponents of stability worked largely with isolates from the US (where rice was introduced only a few hundred years earlier) and that had been in culture for a number of years.
Somewhere in the middle emerged the proposition that the key point for breeding was being overlooked. Blast populations are very diverse, regardless of the mechanisms (genetic or otherwise) that generated the diversity, and the design of most breeding programs is such that a blast-resistant variety is simply not exposed to pathogenic variants that it would likely encounter under production conditions (2). In other words, real-world rice varieties would be exposed to populations of the pathogen, not just one or two races. To address this limitation, a breeding program was established in a rice blast hot-spot region of Eastern Colombia (Santa Rosa) in which segregating progeny of crosses were evaluated under very high natural disease pressure during their entire vegetative and reproductive phases and in every generation from the F2 to the F6, or later (6) (Figure 4). This yielded durably resistant varieties (5), with one (Oryzica Llanos 5) remaining resistant in eastern Colombia for the past 10 years of commercial cultivation, and showing high levels of resistance in trials around the world (R. Zeigler, personal observations). This success notwithstanding, failure to understand how such resistance was achieved made it difficult to replicate in other areas, particularly in those areas where disease pressure was less reliable.
The discovery of a neutral repetitive DNA sequence, MGR (for Magnaporthe grisea repeat) in the rice blast pathogen in the late 1980s provided a means of analyzing populations independently of the pathogenicity of the constituent isolates (7). The similarity of the MGR "fingerprints" (Figures 5 and 6) generated by analyzing the DNA of different isolates permitted an estimate of their relatedness. Initial analysis of archival US P. grisea isolates revealed a direct relationship between fingerprint type (subsequently referred to as lineages) and pathogenic races (11). Application of this analytical tool to the Santa Rosa population (12) yielded a less direct, but intriguing, relationship between lineage and race: Sets of closely related races fall within a single lineage and the race constitution of lineages differed. Furthermore, in what had been described as an extremely race-diverse population, all isolates could be grouped into only six lineages. This led to the suggestion that rice breeding could focus on selecting for cultivars that combined resistance that was effective against the virulence spectrum of all lineages in a target population.
This breeding approach, referred to as "lineage exclusion" (24), assumes that P. grisea populations are comprised of a few number of discrete lineages and that these lineages have different and stable virulence spectra. Zeigler et al. (23) tested these assumptions in two populations from blast resistance screening nurseries in the Philippines. They found that, like in Colombia, there were relatively few lineages comprising the populations (3). Analysis of lineage virulence spectra (i.e., the virulence of isolates on sets of isolates with known and different resistance) revealed that they were indeed different (23). "Composite pathotypes" could be created for a lineage by considering any compatibility within a lineage as reflecting the virulence capacity of that lineage. Comparing the composite pathotypes of all the lineages of a population could predict what combination of resistance would be effective across the entire population. In the case of the Philippines, a combination of resistance genes Pi-1 and Pi Z5 (Pi-2) should yield resistance effective across all lineages (Table 1).
A similar analysis in Santa Rosa also predicted that the same two genes should yield broad-spectrum resistance. This was tested by crossing two sources of resistance and then evaluating the progeny in the field (exposing them to a diverse, well-characterized population) and in the greenhouse (exposing them to isolates representing the full virulence spectrum in all lineages in the population). As expected, progeny resulted with full spectrum resistance in both greenhouse and field evaluations (Table 2). Based on this positive result, parents in crosses for blast resistance in Santa Rosa have recently been selected to combine complementary resistance. This has yielded an significant increase in the efficiency of the breeding programs (Table 3).
How effective can lineage exclusion be as a breeding tool for obtaining durable blast resistance world-wide? A few critical issues suggest that with present technology, all areas may not be suitable for its adoption. The situations in the Philippines and Santa Rosa may be somewhat atypical in that these populations are from areas where modern varieties have been grown and, because of a bottleneck effect of earlier deployed blast resistance, the pathogen population may be much simpler than those populations in other rice-growing regions. i.e. if populations are very complex it could be practically impossible to characterize the virulence spectra of all lineages. Furthermore, for lineage exclusion to yield durable resistance, lineages should be genetically isolated from one another so that virulence genes cannot be exchanged among lineages.
A population analysis of P. grisea from a traditional rice-growing area of northeast Thailand revealed a very complex population: 49 lineages were identified from 527 isolates, and most were represented by only one or a few isolates (14). No obvious relationships between pathotype and lineage was discerned within these samples using either lines near-isogenic for resistance genes or cultivars with known resistance. Very high lineage diversity was also observed in the Indian Himalayas (8) and very high pathotypic diversity was observed in the Himalayan Kingdom of Bhutan, although the corresponding lineage data are sketchy (18,19). It would be impossible to determine the virulence spectrum of lineages comprising these populations. First, many lineages are represented by only one isolate, so there is no way to determine the virulence spectrum of the lineage. Second, there are so many lineages that the logistics of determining virulence spectrum greatly exceed the capacities and resources of the breeding programs working in these areas. These problems notwithstanding, the analysis of the NE Thailand population revealed the same complementary effectiveness of resistance genes Pi 1 and Pi z5.
An important assumption of the lineage exclusion approach is that there is no gene flow across or genetic recombination among lineages. Several lines of evidence suggest that this may not be the case in some areas. Reports of sexually fertile field isolates from India (8,20), China (4), and Thailand (15) indicate that the capacity for sexual recombination exists in nature. Population structure and dynamics of Indian Himalayan populations are consistent with sexual recombination having influenced populations there (8). There is also the possibility that horizontal flow of genes, including those mediating resistance to entire lineages, can occur across lineages via non-sexual, or parasexual, means (21).
Despite indications that there may be very large areas over which a population analysis-based lineage exclusion breeding strategies may not be appropriate, there is ample evidence that population analyses can yield valuable dividends. First, in most cases examination of the virulence spectra of the most common lineages should indicate to breeders which crosses are unlikely to yield durable blast resistance, thus increasing their efficiency. Second, the repeated conclusion that the gene combination Pi 1 and Pi z5 is effective across very different populations suggests there is something fundamentally limiting to P. grisea carrying compatibility to both genes simultaneously. As more blast resistance genes are identified and placed in near-isogenic backgrounds population analyses will enable us to identify other broadly effective gene combinations. Finally, there are large and important rice growing areas where P. grisea populations are relatively simple. These may be where rice has only recently been introduced, or where very large areas have been planted to a few varieties carrying several major resistance genes. The former areas include all of the Americas, Europe, Africa and, probably, Australia. The latter areas may include the extremely important rice growing areas of Southeastern China, Java, the Central Luzon area of the Philippines, parts of the Indo-Gangetic Plain and Central India. Thus, breeding strategies for these areas should be adjusted accordingly.
References Cited
1. Bonman, J.M., Estrada, B.A., Kim, C.K., Ra, D.S., and Lee, E.J. 1991. Assessment of blast disease and yield loss in susceptible and partially resistant rice cultivars in two irrigated lowland environments. Plant Disease 75:462-466.
2. Buddenhagen, I.W. 1983. Breeding strategies for stress and disease resistance in developing countries. Annual Review of Phytopathology 21:385-409.
3. Chen, D., Zeigler, R.S., Leung, H., and Nelson, R.J. 1995. Population structure of Pyricularia grisea at two screening sites in the Philippines. Phytopathology 85:1011-1019.
4. Chengyun, L., Jiarui, L., Rui, S., and Yoshikatsu, F. 1992. Cross-fertility of rice blast fungus Pyricularia orzya. Southwest Journal of Agricultural Sciences 5:53-58.
5. Correa-Victoria, F.J., Zeigler, R.S. 1995. Stability of partial and complete resistance in Rice to Pyricularia grisea under rainfed upland conditions in eastern Colombia. Phytopathology 85:977-982.
6. Correa-Victoria, F.J., and Zeigler, R.S. 1993. Pathogenic variability in Pyricularia grisea at a rice blast "hot spot" breeding site in eastern Columbia. Plant Disease 77:1029-1035.
7. Hamer, J.E., Farral, L., Orbach, M.J., Valent, B., and Chumley, F.G. 1989a. Host species-specific conservation of a family of repeated DNA sequences in the genome of a fungal plant pathogen. Proc. Natl. Acad. Sci. (USA) 86:9981-9985.
8. Kumar, J., Nelson, R.J., and Zeigler, R.S. 1999. Population structure and dynamics of Magnaporthe grisea in the Indian Himalayas. Genetics 152:971-984.
9. Latterell, F.M. 1971. Phenotypic stability of pathogenic races of Pyricularia oryza, and its implications for breeding of blast resistant rice varieties. Pages 199-234 in: Proceedings, Seminar on Horizontal Resistance to Blast Disease of Rice, Centro International de Agricultura Tropical (CIAT), Cali, Colombia.
10. Latterell, F.M., and Rossi, A.E. 1986. Longevity and pathogenic stability of Pyricularia oryza. Phytopathology 76:231-235.
11. Levy, M., Romao, J., Marchetti, M.A., and Hamer, J.E. 1991. DNA fingerprinting with dispersed repeated sequence resolves pathotype diversity in the rice blast fungus. Plant Cell 3:95-102.
12. Levy, M., Correa, F.J., Zeigler, R.S., Xu, S. and Hamer J.E. 1993. Genetic diversity of the rice blast fungus in a disease nursery in Colombia. Phytopathology 83:1427-1433.
13. Marchetti, M.A. 1983. Dilatory resistance to rice blast in USA rice. Phytopathology 73:645-649.
14. Mekwatanakarn, P., Kositratana, W., Levy, M., and Zeigler, R.S. 2000. Pathotype and avirulence gene diversity of Pyricularia grisea in Thailand as determined by rice lines near-isogenic for major resistance genes. Plant Disease 84:60-70.
15. Mekwatanakarn, P., Kositratana, W., Phromraksa, T., and Zeigler, R.S. 1999. Sexually fertile Magnaporthe grisea rice pathogens in Thailand. Plant Disease 83:939-943.
16. Ou, S.H. 1979. Breeding rice for resistance to blast--A critical review. Pages 81-137 in Proceedings, Rice Blast Workshop. International Rice Research Institute, PO Box 933, Manila, Philippines.
17. Ou, S.H. 1985. Rice Diseases. 2nd Edition, Commonwealth Mycological Institute, Kew, UK, 380 pp.
18. Thinlay, Finckh, M.R., Bordeos, A.C. and Zeigler, R.S. 2000. Effects and possible causes of an unprecedented rice blast epidemic in the traditional farming system of Bhutan. Agric. Ecosyst. Environ. (In Press)
19. Thinlay, Zeigler, R.S., and Finckh, M.R. 2000. Pathogenic variability of Pyricularia grisea from the high- and mid-elevation zones of Bhutan. Phytopathology 90:621-628.
20. Zeigler, R.S. 1998. Recombination in Magnaprthe grisea. Annual Review of Phytopathology 36:249-276.
21. Zeigler, R.S., Scott, R.P., Leung, H., Bordeos, A.A., Kumar, J., and Nelson, R.J. 1997. Evidence of parasexual exchange of DNA in the rice blast fungus challenges its exclusive clonality. Phytopathology 87:284-294.
22. Zeigler, R. S., Teng, P. S., Leong, S. A. 1994. Rice Blast Disease. Commonwealth Agricultural Bureaux, Wallingford, UK. 626 pp.
23. Zeigler, R.S., Cuoc, L.X., Scott, R.P., Bernardo, M.A., Chen, D.H., Valent, B., and Nelson, R.J. 1995. The relationship between lineage and virulence in Pyricularia grisea in the Philippines. Phytopathology 85:443-451.
24. Zeigler, R.S., Tohme, J., Nelson, R. J., Levy, M., Correa, F. J. 1994. Lineage exclusion: A proposal for linking blast population analysis to resistance breeding. pp. 267-292 in Rice Blast Disease, R. S. Zeigler, P.S. Teng, S. A. Leong (eds.) Commonwealth Agricultural Bureaux, Walllingford, UK.
RETURN TO APSnet FEATURE STORY
