The final result of the interaction that occurs among all of the evolutionary forces is the genetic structure of a pathogen population. Genetic structure refers to the amount and distribution of genetic variation within and among populations. This is due to the summed effect of all evolutionary forces that have acted on a population over time. The evolutionary forces of mutation, genetic drift, gene/genotype flow, reproductive/mating systems, and selection operate together to affect the genetic structure of populations.
The interactions among evolutionary forces are best illustrated by using several examples from the plant pathology literature:
Though many forces may interact to determine pathogen evolution, the interaction between mutation and selection remains the dominant paradigm to explain the evolution of plant pathogens based on experience with boom-and-bust cycles with cereal rusts in North America, Europe, and Australia. In a boom-and-bust cycle, a resistant cultivar with single, major resistance gene is introduced into an agroecosystem to control a plant disease. If the resistant cultivar has good agronomic characters and is widely accepted by farmers because it is disease-resistant, the cultivar spreads and is planted over a large area. This is the "boom" part of the cycle, characterized by an increase in the area planted to the resistance gene (Figure 32). In the pathogen population exposed to this resistance gene, a mutation from avirulence to virulence occurs. The mutation could be the loss of the elicitor recognized by the resistance gene. Selection imposed by the resistant cultivar increases the frequency of pathogen strains with the virulence mutation, usually in a time-lagged manner that shadows the increase in frequency of the resistance gene (Figure 32). The virulent pathotypes spread (by gene or genotype flow) and infect all fields with the resistant cultivar, causing an epidemic and leading to a loss of effectiveness of the resistance gene. Because the resistance is "broken", farmers stop planting the resistant cultivar and the corresponding resistance gene decreases in frequency. This is the "bust" part of the cycle (Figure 32). The cycle begins again with the introduction of a new resistant cultivar.
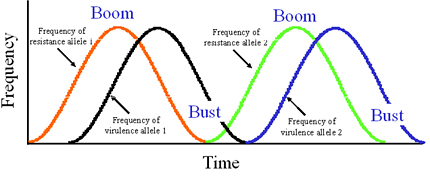 |
| Figure 32. A phase-diagram representation of the boom-and-bust cycle showing the phase lag oscillation in frequencies of major resistance alleles in the host population and corresponding virulence alleles in the pathogen population. Click here to see an enlarged view of this figure. |
The boom and bust cycle is colorfully described in the following excerpt from EC Stakman’s 1957 manuscript. He shows that gene flow is involved in the Puccinia pathway.
|
Excerpt from Stakman's Manuscript
Following the terrible epidemic of 1916, rust-resistant durum wheats, from which macaroni is made, were substituted extensively for bread wheats in the spring wheat region of the United States, but within a few years rust races appeared to which they were completely susceptible. Kanred, a good winter wheat selected in Kansas, appeared to be immune from rust, but almost as soon as it was distributed rust races were found to which it was completely susceptible. In 1926 the rust-resistant hybrid variety Ceres, a good spring wheat, was distributed and soon became by far the most popular variety in the spring wheat region of Minnesota, the Dakotas, eastern Montana, and adjacent Canada. Ceres seemed to have solved the stem rust problem in spring wheat. But in 1935 it was suddenly and tragically ruined in a terrific epidemic caused principally by race 56 of stem rust. This race had first been isolated from barberry in Iowa in 1928, two years after Ceres was first distributed, and by 1934 it was the most prevalent race in the United States. In 1935 race 56 almost literally exploded and virtually ended the career of Ceres wheat. Many other varieties, however, had been in the making, among them Thatcher, which resulted from crosses involving three parents. It had been severely tested against all known races of stem rust prior to its first distribution in 1934. It came unscathed through the epidemic of 1935 and another severe epidemic in 1937. It was extremely susceptible to orange leaf rust and to Fusarial head blight, however, so that it could not be grown successfully in the more humid parts of the spring wheat area where these diseases are most prevalent and destructive. It persisted, however, in the drier areas of Western Canada, where it still produces well on millions of acres.
Another series of varieties were soon released that had the "Hope type" of resistance. The variety Hope resulted from a cross between Jaroslav emmer, which had no virtues except rust resistance, and Marquis wheat, which had all the virtues except rust resistance. Hope did not have the qualities needed in a commercial bread wheat, but it was widely and successfully hybridized with good bread wheats. Similarly, highly resistant durums had been produced by crossing good macaroni varieties with Vernal emmer. Thus the era of "Hope resistance" began. Again it looked as if stem rust was under control in the spring wheat area.
From 1938 to 1949, inclusive, there were no epidemics of stem rust in the spring wheat area of the United States and Canada. As barberry eradication progressed, the number of prevalent rust races decreased. Only four were prevalent enough to be important during this period, and the varieties grown were resistant to all of them. But many different races were being found on barberries in eastern United States. Among them was the very virulent race 15B, to which all of the resistant varieties of bread wheat and durum then grown were susceptible. The question as to whether this very dangerous race would ever become widespread and prevalent was answered suddenly and dramatically in 1950. An unusual sequence of wind and weather conditions enabled the improbable to happen: race 15B spread over most of North America in a single season. A few spores apparently were blown into the Gulf States early in the spring, then 15B appeared sparingly in Texas and gradually spread northward, gathering momentum as it went onward to southern Canada. A very late crop season and a late and wet fall in the north enabled the rust to produce countless billions of spores on the very late wheat and on wild grasses, especially wild barley. Now the question arose as to whether winter would descend on the rust and freeze it to death before favorable winds could carry spores to the far south, where they could establish the rust for the winter. Winds did their worst; they carried a heavy cargo of spores far southward into Mexico, where the rust became established, survived the winter, and was ready to reinfect wheat fields to the northward in the spring. Thur the most virulent race of wheat stem rust ever found in North America spread over most of the continent and became independent of barberry in a single year. The consequences were tragic. Race 15B ruined thousands of acres of the most resistant wheat that had been laboriously developed in Mexico, it caused heavy local damage to all resistant varieties in northern United States and Canada, and in 1953 and 1954 it ruined the durum wheat crop of the United States and caused extensive damage to hitherto resistant bread wheats. |
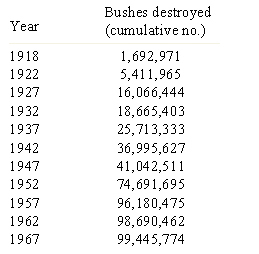
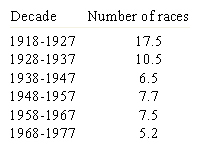
Recombination and selection affected the diversity in Puccinia graminis f. sp. tritici pathotypes in North America. Before barberry eradication (~ 500 million barberry bushes were removed over 50 years in North America), hundreds or thousands of pathotypes of the stem rust pathogen P. graminis f. sp. tritici existed as a result of recombination on the alternate host. The sexual stage of this pathogen occurs only on the barberry host. By 1950, only four races were found in abundance (Roelfs, 1982). After removal of alternate hosts (Table 7), the sexual cycle no longer occurred and efficient recombination through meiosis disappeared as well, leading to a reduction in the genotypic diversity in the rust population (Table 8). Resistance gene pyramids began working in North America because the pathogen population stabilized into a limited number of clonal lineages. The only source of diversity in the rust population became mutation within clonal lineages. Recombination was no longer available as a means of assembling new virulence allele combinations.
| Table 7. Cumulative number of barberry bushes destroyed at the end of 5-year periods in the original 13 barberry eradication states (Roelfs 1982). |
 |
|
| Table 8. Average number of wheat stem rust races found per year in each decade since 1918 in the United States (Roelfs 1982). |
 |
The interaction between drift, genotype flow, and selection is described for Puccinia graminis f. sp. tritici in Australia by Burdon and Silk (1997) and is summarized in Figure 33. The original introduction of the pathogen occured during the European settlement of Australia in the 1780s. A few strains of the pathogen were introduced into Australia, leading to a founder effect. No barberries were present in Australia, so no sexual cycle existed and genotypic diversity was limited. As a result, the pathogen population was made up of several clonal lineages, that acquired mutations from avirulence to virulence within each clonal lineage. One clonal lineage (called race 126) dominated Australian wheat fields from 1921-1954. In 1954, a new race emerged (race 21), apparently carried on the jet stream from East Africa (an example of genotype flow). Within 3 years, race 21 completely replaced race 126. Race 21 had new virulence alleles, new isozyme markers, and new DNA polymorphisms. Long-distance gene flow could explain the origins of two other dominant clonal lineages that emerged in the 1970s in Australia (clones 326 and 194). This appears to be a good example of genotype replacement, where new, more-fit genotypes replaced the previously existing genotypes as a result of directional selection.
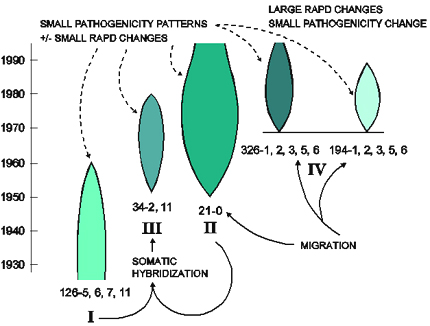 |
| Figure 33. Diagrammatic representation of the origin and structure of the Australian Puccinia graminis f. sp. tritici population. I, II, II, and IV refer to separate lineages within the population. The details are discussed in the text. Pathotypes 126-5,6,7,11; 21-0; and 34-2,11 are the founders of lineages I, II, and III, respectively. Pathotypes 194-1,2,3,5,6, and 326-1,2,3,5,6 appeared within 9 months of each other and are assumed to have a common, although unexplained origin. The historical duration of each lineage is represented diagrammatically by the shaded areas, the size of which provides a general guide to the relative importance of each. Solid arrows indicate the origin of lineages, dashed arrows indicate the associations between pathogenicity and molecular markers. Click here for an enlarged view of this figure. |
The interaction between selection and genotype flow is best illustrated by the example of Fusarium wilt of melon and other Fusarium wilts. Fusarium oxysporum is endemic to soils everywhere. Natural populations are aggressive colonizers of the root cortex, but usually do not cause disease. Wild-type isolates appear to be true saprophytes that do not need to kill plants to persist in the soil. Populations of the fungus that are pathogenic on plants appear to be strictly asexual. Formae speciales that infect agricultural and horticultural crops appear everywhere where intensive monocultures of crops are grown. Where do all of these pathogenic Fusarium oxysporum formae speciales come from? A model to explain this process is shown in Figure 34.
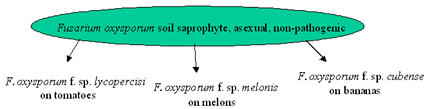 |
| Figure 34. A model to explain the emergence of Fusarium oxysporum wilts in agricultural ecosystems. Monocultures exert selection on the non-pathogenic indigenous Fusarium oxysporum population, causing an increase in the frequency of rare mutants that are pathogenic to the monoculture crop. Genotype flow moves the pathogenic mutants to new locations, leading to an epidemic. Different monocultures select for different F. oxysporum formae speciales from non-pathogenic ancestral populations. |
F. oxysporum pathogens within the same formae specialis can be differentiated into pathotypes according to major resistance genes deployed in host cultivars. Clones of the fungus can be differentiated using vegetative compatibility groups (VCGs), isozymes, or DNA-based markers. It has been observed several times that the same race occurs in different genetic backgrounds, meaning that the same race evolved several times completely independently (Gordon and Martyn 1997). There also are several examples where different races occur in the same genetic background, providing evidence that selection occurred for new mutations from avirulence to virulence in the same clonal lineage (Figure 35).
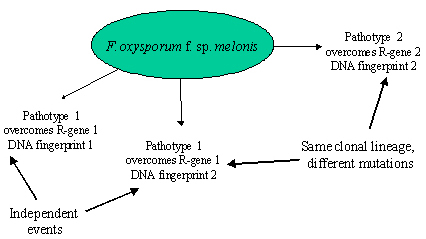 |
| Figure 35. DNA fingerprinting methods made it possible to compare the genetic background of different pathotypes and infer genetic similarities. In the melon wilt pathogen, some strains sharing the same pathotype have different DNA fingerprints, indicating that these strains emerged de novo independently of each other. Other strains with different pathotypes share the same DNA fingerprint, indicating different mutations to virulence in the same clonal lineage (Gordon and Martyn 1997). |
A population genetic interpretation of these observations is that a new pathogenic strain (formae specialis) emerges due to strong selection imposed by an agroecosystem dominated by monoculture, where a uniform host population presents a new niche that can be colonized by any rare mutant pathogenic strain that exists in the highly diverse F. oxysporum population occurring naturally in the soil. This pathogenic strain (or strains) is (are) then moved to new fields, and often around the world on infected or infested plants or soil. The same forma specialis can originate independently in different agroecosystems, generating wilt strains with independent genetic backgrounds. As an example, F. oxysporum f. sp. cubense (causing Panama Disease on bananas) appears polyphyletic. Two clonal lineages with independent origins have moved around the world (Koenig et al. 1997).
This interpretation is consistent with the clonal genetic structure found for all of the Fusarium oxysporum formae speciales characterized thus far, including wilts on carnation (Migheli et al. 1998), coca (Nelson et al. 1997), oil palm (Mouyna et al. 1996), cyclamen (Woudt et al. 1995), tomato (Bao et al. 2002), lily (Baayen et al. 1998), chickpeas (Jimenez-Gasco et al. 2002), and date palm (Plyler et al. 2000). Thus it appears that Fusarium oxysporum wilts will emerge spontaneously wherever monocultures dominate and the potential for genotype flow exists.
Mycosphaerella graminicola (anamorph Septoria tritici) on wheat presents a good example of the interaction between recombination and gene flow. RFLP markers have shown that populations around the world have a high degree of genotypic diversity (Figure 36), a low degree of clonality, and random associations among loci, as expected for random-mating populations that exhibit a high degree of gene flow (Linde et al. 2002; Zhan et al. 2003). The sexual cycle appears to occur in field populations around the world. Neutral RFLP markers show a high degree of similarity among field populations around the world (Figure 37). The same RFLP alleles are present at nearly identical frequencies in populations collected from Oregon and Israel, but no genotypes are shared among field populations. A field experiment provided evidence that the balance between sexual reproduction and immigration changed over the course of the growing season. At the beginning of the season, most genetic diversity came from immigration as a result of ascospores arriving from distant source populations. But by the end of the growing season, most of the genotypic diversity came from recombination among isolates within the same field (Zhan et al. 1998).
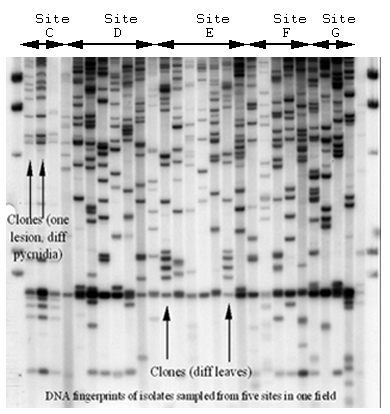 |
| Figure 36. DNA fingerprints of Mycosphaerella graminicola isolates sampled from five sites separated by 10 m in one field. Clonality exists over spatial scales of a few meters within a field, but no clones are shared between fields. |
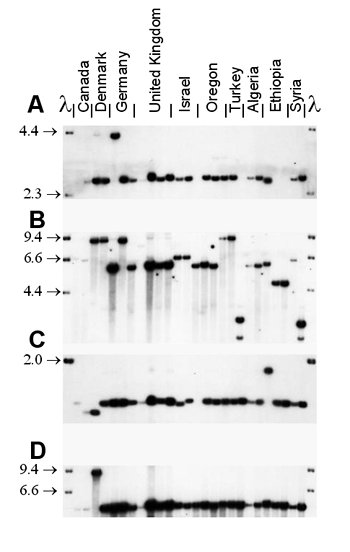 |
| Figure 37. An autoradiogram showing conservation of RFLP alleles in Mycosphaerella graminicola populations from around the world. M. graminicola DNA was digested with Pst1 and lambda DNA was digested with HindIII. A) Locus pSTL10. B) Locus pSTL31. C) Locus pSTS14. D) Locus pSTS192A. |
Go to Knowledge Test for Interactions/Genetic Structure
Go to References
Next Section
