Plant pathologists experienced pathogen evolution firsthand many times as pathogen populations evolved to overcome resistance genes or became resistant to fungicides or antibiotics. In this final section we will use principles of population genetics to evaluate the risk of pathogen evolution. Our hypothesis is that the pathogens with the greatest evolutionary potential offer the greatest "risk" of overcoming major resistance genes or evolving to counteract other control methods such as applications of fungicides or antibiotics.
Pathogens with high mutation rates present a greater risk than pathogens with low mutation rates because a high mutation rate increases the likelihood that the mutation from avirulence to virulence (or from fungicide sensitivity to fungicide resistance) will be present in a pathogen population. Mutation rates are generally low, though they can differ among loci and pathogens. Pathogen populations with active transposable elements may pose a greater risk than populations without active transposable elements. But as shown earlier, a mutation operating in isolation from other evolutionary forces is unlikely to occur at a high enough rate to produce detectable changes in allele frequencies.
Pathogens with large population sizes have more evolutionary potential than pathogens with small population sizes because more mutant alleles are present in larger populations. Pathogens that undergo regular, severe reductions in population size, e.g. as a result of crop rotations or annual climatic extremes that kill the majority of individuals, are less diverse as a result of genetic drift and slower to adapt than populations that maintain a high population size year round. Two simple ways to minimize pathogen population size are by using regular crop rotations and by avoiding the cultivation of extremely susceptible cultivars that allow pathogen population sizes to increase explosively.
Pathogens with high gene flow pose a greater risk than pathogens with low gene flow for two reasons. 1) Populations with high gene flow are larger, and thus maintain more alleles. 2) Pathogen populations with high gene flow are more likely to transmit virulent or fungicide resistant mutants across a large geographical area. Gene flow involving asexual propagules (genotype flow) poses greater risks than movement of sexual propagules (gene flow) because the asexual propagule represents a linked package of coadapted genes that was pre-selected for a high level of fitness in the crop environment where it originated. Sexual propagules (e.g. ascospores) represent new combinations of alleles that have not yet been tested against the environment where they will land. In this case, the disease management objective is to limit the extent of gene flow among pathogen populations. While we cannot affect "natural" dispersal of pathogen propagules by wind, water, or insects, we can limit the potential for long-distance dispersal aided by man, including movement of infected plant material, soil, or contaminated equipment among otherwise isolated pathogen populations. We can also attempt to eliminate "living bridges" of susceptible host materials that can act as stepping stones between populations, or plant strips of resistant plants (or non-host plants) to act as barriers between adjacent susceptible host populations.
Pathogens that undergo regular recombination (this can include processes distinct from meiosis such as bacterial conjugation, recombination between viral genomes in plants with mixed infections, and hyphal anastomosis and/or parasexual recombination in fungi) pose higher risks than pathogens that undergo no or little recombination. Recombination allows new combinations of alleles to come together and be tested against new environments. A pathogen that undergoes regular recombination can put together new combinations of mutant alleles (e.g. virulence alleles) as rapidly as breeders can recombine major resistance genes. For this reason, resistance gene pyramids may not be an effective long-term breeding strategy against pathogens that undergo regular recombination.
Pathogens with mixed reproductive systems that include both sexual and asexual reproduction pose the highest risk of evolution because they receive benefits from both mechanisms of reproduction. Sexual recombination allows many new combinations of alleles to come together and then be tested in the local environment. Asexual reproduction allows the most fit genotype to reproduce as a clone, keeping together a fit combination of alleles and making it possible for this allele combination to become widely distributed if the asexual propagules are dispersed over long distances.
Pathogen populations that are exposed to strong, directional selection over many generations pose a greater risk than populations that are exposed to weaker selection as a result of partial resistance or to disruptive selection resulting from temporal or spatial patterning of the selective force. Selection is the evolutionary force that is most easily manipulated by humans, and thus offers the most practical point for intervention in the evolutionary process. Agroecosystems based on widespread deployment of single, major resistance genes (genetically uniform monoculture) place strong directional selection on the pathogen population. Agroecosystems that deploy major resistance genes in mixtures, or in rotations through time and space will reduce the efficiency of selection, or impose stabilizing or disruptive selection that can slow the rate of increase in the frequency of virulent mutants
The contributions of each evolutionary force to the assessment of risk are summarized in Table 9.
Table 9. Extremes of evolutionary risk posed by plant pathogens and example factors that affect risks (from McDonald and Linde 2002).
|
Highest risk of evolution to overcome control method |
Lowest risk of evolution to overcome control method |
|
High mutation rate.
Transposable elements active. |
Low mutation rate.
No transposons. |
|
Large population sizes. Large overseasoning population. Local extinction rare. No genetic drift, no alleles lost. |
Small population sizes. No overseasoning propagules. Extinction of local populations common. Significant genetic drift, alleles lost. |
|
High gene/genotype flow. Asexual propagules dispersed by air over long distances. Human-mediated long distance movement common. |
Low gene/genotype flow. Asexual propagules soil-borne. Quarantines effective. |
|
Mixed reproduction system. Annual sexual outcrossing and asexual propagules produced. |
Asexual reproduction system. Only asexual propagules produced. |
|
Efficient directional selection. R-gene deployed in genetically uniform monoculture. R-gene deployed continuously over large area. |
Disruptive selection. R-genes deployed in mixtures/multinlines. R-genes deployed as rotations in time or space. |
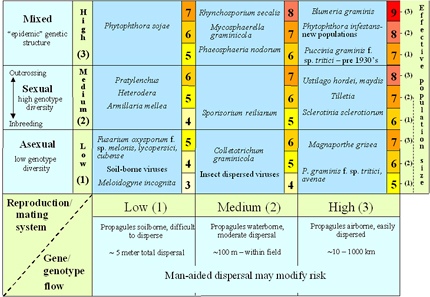
Figure 38 combines information on the evolutionary risks due to differences in reproduction/mating system, gene flow and effective population size and places different pathogens into separate risk categories according to what is known about their population genetics. Mutation rate was not included in this risk assessment under the assumption that mutation rates are low and relatively constant across all pathogens, so the differences in this parameter for different pathogens on average are expected to be relatively small. Selection was not included under the assumption that selection is likely to be efficient in the monocultures that dominate modern agricultural ecosystems so that mutants will increase in frequency quickly if a selective influence is present. Selection risk can be adjusted by modifying the environment through different strategies of resistance gene deployment, such as gene rotations or mixtures (Figure 39). It is important to recognize that these proposed risk categories need to be adjusted on a case-by-case basis as a result of anthropogenic activities. For example, gene flow may be increased beyond the normal biological limits of spore dispersal by movement of inoculum through international commerce and travel. Any activities that reduce the effectiveness of quarantines, such as smuggling or war, may lead to an increase in gene flow. Similarly, the amount of sexual reproduction may be affected by removal of alternate hosts or by changes in cultivation or sanitation practices.
The risk values presented in Figure 38 are on a 3-9 scale. These numbers are unitless and have no specific biological meaning. The numbers provide a relative ranking of the evolutionary potential inherent in different pathogen life histories. The present ranking system assumes that reproduction/mating system, gene flow and effective population size affect evolutionary potential equally, and that these effects are additive. Figure 38 should be considered a preliminary guide for assigning risk.
 |
|
Figure 38. A risk assessment chart that combines estimates of gene flow, effective population size, and reproduction/mating system to place pathogens into different categories of evolutionary potential (from McDonald and Linde 2002). Click here to see an enlarged view of this figure.
|
 |
|
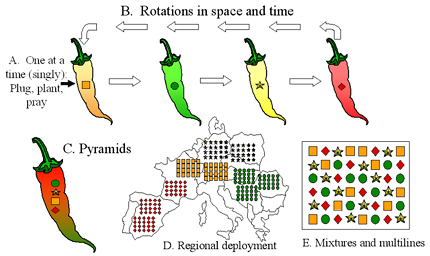
Figure 39. Deployment strategies for major resistance genes. A) The traditional "plug, plant, and pray" approach of deploying single R genes one at a time in a sequence that matches the boom and bust cycle. B) Rotations of R genes in time or space. Each R gene is deployed over a limited number of years or limited area, and is withdrawn before the corresponding virulence allele achieves a high frequency in the pathogen population. C) R gene pyramid. All R genes are placed together in one plant genotype. D) Regional deployment. Different R genes are grown in different regions. This can be part of a gene rotation as indicated in B. E) Cultivar mixtures and multilines. Individual R genes are grown as an intimate mixture in the same field. Click here to see an enlarged view of this figure. |
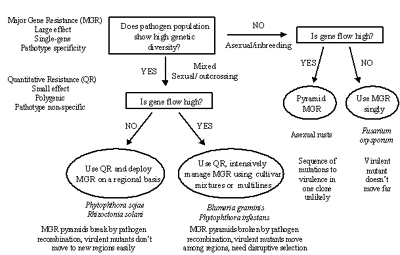
Figure 40 presents a guide for deployment of resistance genes based on knowledge of population genetics of pathogens and hosts. It assumes that major-gene resistance (abbreviated MGR) is based on the receptor-elicitor model of the gene-for-gene interaction and that minor-gene resistance (abbreviated QR for "quantitative resistance") is a quantitative character based on several unlinked genes that show equal and additive effects. The diagram assumes that both types of resistance are available to the plant breeder, and that major-gene resistance is preferred because it is easier to recognize and is more easily introgressed into good agronomic types. To keep the diagram as simple as possible, the reproduction/mating systems of the pathogen are assumed to be either asexual/inbreeding, or mixed/outcrossing. Gene/Genotype flow is assumed to be either high or low. This diagram can be extended with many additional branch points to encompass the full range of possible interactions among evolutionary forces and potential pathogen population genetic structures. Effects of different mutation rates, population sizes, or the full range of reproduction/mating systems were not included. The diagram was kept simple in an effort to illustrate general principles. Click here to see an enlarged view of this figure.
The outcome of the decision diagram is a general recommendation for choosing the type of resistance and the optimal deployment method, (with the aim of maximizing the useful life span of the resistance). This diagram should be considered in the appropriate context in a resistance-breeding program, as an aid to assist with decision making. It is not intended as an authoritative guide that is certain to lead to durable resistance.
Go to References
Next Section
