Randy C. Ploetz
University of Florida
Tropical Research & Education Center
18905 SW 280th Street
Homestead, FL 33031-3314 USA
rcp@ifas.ufl.edu
Ploetz, R.C. 2005. Panama Disease: An Old Nemesis Rears its Ugly Head Part 2: The Cavendish Era and Beyond. Online. APSnet Features. doi: 10.1094/APSnetFeature-2005-1005
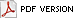
The start of the banana export trades and the influence that Panama disease had on their development was discussed in the August feature article. The second part of this review begins with a brief introduction to banana taxonomy and diversity. Banana is a highly variable crop, and Panama disease impacts many of the different types that are grown.
The current impact of Panama disease is summarized with special reference to tropical race 4 (TR4). TR4 affects cultivars that produce more than 80% of the world’s bananas, including the important Cavendish and plantain subgroups. If TR4 were to spread outside its current, limited range, its potential impact would be greater than that caused by race 1 during the ‘Gros Michel’ era (see August APSnet Feature).
Also discussed is phylogenetic research on the causal fungus, Fusarium oxysporum f. sp. cubense (FOC). Recent work on formae speciales (ff. spp.) of F. oxysporum has provided useful insight into the origins and relatedness of these important plant pathogens. Since most of the ff. spp. affect closely related taxa it had been assumed that members of each were also closely related (13). Surprisingly, recent work indicates that this is not always the case. Many ff. spp. have been shown to have evolutionarily diverse backgrounds (20). These results have significant implications for the development of resistant host genotypes and, for FOC, make the challenging job of improving banana more difficult (43).
This article ends with information on banana improvement. Traditional breeding programs face several obstacles. These difficulties, the limited success with some breeding targets (especially the export dessert bananas), and the serious threat that diseases pose to current production have led the popular press to write that banana will become extinct in the near future (15,23). Although these articles exaggerate the magnitude and ultimate impact of these problems (1,19), they have drawn the general public’s attention to the importance of these threats and to the biotechnological improvements that are envisioned for the crop.
Banana Taxonomy and Diversity
Bananas and plantains (a type of banana) are monocotyledenous herbs from southern Asia (16). With few exceptions, the cultivars are parthenocarpic landraces that are propagated vegetatively (traditionally with rhizomes, and more recently with tissue-culture plantlets). They are complex diploid, triploid and tetraploid hybrids among subspecies of Musa acuminata, and between M. acuminata and M. balbisiana, both of which are diploid. A shorthand system is used whereby the ploidy and genomic constitution of cultivars is denoted with an A/B lettering system, with "A" referring to M. acuminata and "B" referring to M. balbisiana (40). For example, ‘Gros Michel’ and the Cavendish cultivars are triploid, pure M. acuminata and thus AAA, whereas the true plantains are triploid, 2/3 M. acuminata, 1/3 M. balbisiana and AAB. The Linnaean binomials M. paradisiaca (the plantains) and M. sapientum (the sweet dessert bananas, of which ‘Silk’ AAB is the type cultivar) are invalid since they refer to interspecific hybrids. The genomic constitutions of different banana cultivars are indicated with this lettering system where they first appear below.
For those of us who grew up thinking of a banana as what we sliced on our breakfast cereal it may come as a surprise that there are many different kinds of this fruit. About 1,000 cultivars in 50 different subgroups are recognized (11). Bananas come in diverse shapes, sizes and colors (Fig. 1). They are used in many ways: brewed for beer, wine and gin (see 8); baked, boiled and fried; and, of course, eaten ripe. In addition, the male buds are used as vegetables, the leaves are used as wrapping material, the pseudostems are used to make paper and as animal feed, and the sap can be used as an indelible dye.
| |

Fig. 1. Bunches of different banana clones at the Fundación Hondureña de Investigación Agrícola (FHIA) in Honduras. Note the diversity of bunch and fruit phenotypes (photo courtesy of R. C. Ploetz). |
|
Only a small number of the known banana cultivars produce most of these fruit worldwide (Fig. 2) (11). The Cavendish subgroup is most significant. The export trades in dessert bananas, which account for 13% of global production, depend almost entirely on these clones. However, locally consumed Cavendish fruit are more important and are responsible for an additional 28% (Fig. 3). Thus, 41% of the world total comes from only one of the 50 subgroups that are known. The plantains, next in importance, are responsible for 21%, and global figures are rounded out by heterogeneous collections of cooking cultivars (these are distinct from the plantains and total 24%) and dessert bananas (14%). Overall, a remarkably small portion of the crop’s diversity is significant in world production. The reliance on such a limited number of clones has particular relevance to the threat that is posed by TR4 (30,31,35).
| |
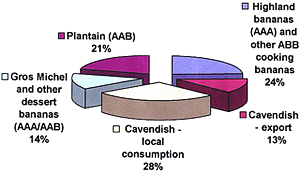
Fig. 2. Relative global production of fruit for different subgroups and types of banana (data are from ref. 11). |
|
| |

Fig. 3. Bunches of ‘Dwarf Cavendish’ AAA for sale at a roadside stand in East Africa. Cavendish fruit are most important in the export trades, but more than twice as many of these fruit are produced for local consumption (photo courtesy of R. C. Ploetz). |
|
Pathogenic Diversity in FOC
FOC affects primarily banana, but other banana relatives are also susceptible. Abacá, M. textilis, source of an important cordage fiber, suffered commercial losses before production ceased in Central America in 1956 (42,48). M. schizocarpa, a parent of rare bananas in Papua New Guinea, is highly susceptible, and several species of Heliconia were reported as hosts in the mid-1900s (49). None of the above species are considered below.
The well-known epidemics of Panama disease on ‘Gros Michel’ tend to obscure its impact outside the export trades and on other banana cultivars (27,29). Diverse clones are attacked, many of which are as susceptible as ‘Gros Michel.’
Three races of FOC are conventionally recognized on banana, 1, 2 and 4 (race 3 was described for the heliconia pathogens). Below are listed notable suscepts of each race.
Race 1. Race 1 was responsible for the epidemics on ‘Gros Michel.’ It also affects ‘I.C.2’ AAAA, ‘Silk,’ ‘Pome’ AAB, ‘Pisang Awak’ ABB and ‘Maqueño’ AAB.

Fig. 4. Fingers of ‘Silk’ AAB from a commercial producer in Venezuela that were purchased in a supermarket in Florida. Production by this firm has decreased markedly due to Panama disease (photo courtesy of R. C. Ploetz). |
|
‘I.C.2’ was a hybrid produced by the first banana-breeding program, located at the International College of Tropical Agriculture in Trinidad (37). Named ‘Golden Beauty’ when it was released in 1928, ‘I.C.2’ was developed as a Panama disease-resistant replacement for ‘Gros Michel.’ Its subsequent susceptibility in Honduras is recognized as a failure to account for pathogenic variation in FOC during the screening process. Stover and Buddenhagen (43) reviewed this general problem and the need for a better understanding of pathogenic variability in FOC during the improvement of this crop.
Due to its exceptional flavor, ‘Silk’ may be the most esteemed of all dessert bananas (Fig. 4). Its extreme susceptibility to race 1 has eliminated it in many areas. The cultivar cannot be produced in much of the Western Hemisphere. Significant commercial production was recently decimated in Venezuela and the cultivar is now uncommon in Brazil, one of the world’s leading banana producers. ‘Pome,’ the most important dessert banana in Brazil (Cavendish is relatively unimportant there), is less susceptible than ‘Silk’ to race 1, but is affected nonetheless.
‘Pisang Awak’ is one of the hardiest and most widely distributed bananas worldwide. Over 70% of the bananas that are produced in Thailand are of this clone (Fig. 5), and it has gained wide acceptance in East Africa due to its use as a beer banana and drought tolerance (Fig. 6). In these and other areas, the clone is very susceptible to race 1.
| |
 |
|
 |
|
| |
Fig. 5. A banana grower in Thailand standing in front of a mat of ‘Pisang Awak’ ABB. More than 70% of the bananas that are produced in Thailand are of this cultivar (photo courtesy of R. C. Ploetz). |
|
Fig. 6. A stand of ‘Pisang Awak’ ABB in a relatively arid area in Malawi. Note that the rest of the vegetation in the area is dead. ‘Pisang Awak’ is one of the most drought hardy bananas, and is especially important in areas with monsoon climates (photo courtesy of R. C. Ploetz). |
|
‘Maqueño’ is a member of the Maia Maoli-Popoulu subgroup, which is most prevalent in the Pacific. ‘Maqueño’ has been used as a female parent in the FHIA breeding program (see below), but is unfortunately very susceptible to race 1. Recently, Panama disease began to move into the Pacific, a region that had been generally free of the disease (27; G. Wall, Univ. Guam, personal communication). As Panama disease spreads in the Pacific it threatens ‘Maqueño’ and other cultivars in this important subgroup.
Race 2. Race 2 affects cooking bananas, especially those in the Bluggoe subgroup ABB. The most important cultivar in this group, ‘Bluggoe,’ was widely planted in Latin America before the spread of race 2 and Moko disease, caused by Ralstonia solanacearum. It is also important, but impacted by race 2, in East Africa (Fig. 7).
| |

Fig. 7. An initial focus of Panama disease in a clump of ‘Bluggoe’ ABB on a river bottom in East Africa. FOC is moved effectively by man in infected banana rhizomes, but is also disseminated in running water (photo courtesy of R. C. Ploetz). |
|
‘Bodles Altafort’ AAAA is a race 1-resistant hybrid between ‘Gros Michel’ and ‘Pisang Lilin’ AA that was developed in Jamaica. It succumbed to race 2 when it was deployed to India (44).
Enset, Ensete ventricosum, is an important food crop for 8 million people in Ethiopia (Fig. 8). It is affected experimentally by race 2 isolates from ‘Bluggoe’ and may be the original, African host of an unusual population of FOC (see below; 31).
| |

Fig. 8. Enset, Ensete ventricossum, in the Misuku Hills of Malawi. Unlike most members of the Musaceae that evolved in Asia, enset is endemic to Africa. It is a major food source for 8 million people in Ethiopia, and may be an uncommon, non-banana host on which FOC evolved (see text) (photo courtesy of R. C. Ploetz). |
|
Race 4. Race 4 affects race 1- and race 2-suscepts, cultivars in the plantain and Cavendish subgroups, and diverse additional cultivars, such as ‘Pisang Mas’ AA (34,44,45).
Although they are usually cooked, the plantains are distinct from other cooking bananas, such as those in the Bluggoe and the Lujugira-Mutika subgroups AAA (aka the East African highland bananas). The plantains are important staple foods for many in West Africa and Latin America (11), areas in which race 4 is fortunately not present.
Until recently, the Cavendish cultivars were affected mainly in the eastern subtropics (26). Losses occurred in subtropical Australia (New South Wales and Queensland), the Canary and Madeira Islands and South Africa (Natal and Transvaal) (34). In these areas race 4 is comprised of isolates in vegetative compatibility group (VCG) 0120-01215.
Cavendish is also affected in Taiwan (10). Previous reports indicated that this was another subtropical example of race 4, even though production areas in Taiwan have relatively warm winters and are technically in the tropics (Kaohsiung, 22°S latitude, is south of the Tropic of Cancer) (44). In hindsight, damage in Taiwan may be due to TR4 (see below).
Cold winter temperatures in the subtropics are thought to predispose Cavendish to race 4. For example, photosynthesis (carbon assimilation, A, as µmol CO2/m2/s) of a Cavendish cultivar was reduced 75% during the winter in Queensland, Australia (17). Although damage occurs on Cavendish in the tropics, it is uncommon and also associated with predisposing factors (26). Wilt pockets occurred in Guadeloupe (17°S) where ash from a volcanic eruption lowered soil pH, and in Jamaica (18°S) in low-lying and poorly drained soils above 700 m in elevation.
Thus, Cavendish succumbed in exceptional situations but performed well in good soils in the lowland tropics. Since these cultivars had resisted Panama disease for decades in the same soils in which ‘Gros Michel’ was devastated, it appeared that the ‘Gros Michel’ populations of FOC were incapable of mutating to virulence on Cavendish (4).
The Special Threat Posed by Tropical Race 4 (TR4)
The confidence that Panama disease would not affect the Cavendish clones was shattered in the early 1990s. New Cavendish plantations began to succumb throughout Southeast Asia, the home of banana. This represented the first time that Cavendish was affected in the tropics in the absence of predisposing factors.
The Southeast Asian isolates are in VCG 01213-01216, a unique population that was originally identified in Taiwan and is now known to define TR4. In addition to Taiwan, VCG 01213-01216 is also found in Australia (Northern Territory), islands in Indonesia (Halmahera, Irian Jaya, Java, Sulawesi, and Sumatra), and peninsular Malaysia (34,38,39).
The role that 01213-01216 plays in the Taiwanese epidemics should be examined. Although the distribution and prevalence of different FOC VCGs in Taiwan has not been determined [three other VCGs exist there, 0120-01215, 0121 and 0123 (34)], the climate and the importance of Panama disease on Cavendish on the island suggest that 01213-01216 may be most significant. A closer examination of the situation could shed light on resistance that has been developed in Taiwan among somaclonal variants of ‘Giant Cavendish,’ the so-called ‘Giant Cavendish’ tissue culture variants (GCTCVs) (10). To date, it is not known which VCGs predominate on Cavendish in Taiwan, or what VCGs impact the GCTCVs (Fig. 9).
|
 Fig. 9. Mike Smith (left) and Ken Pegg (right), of the Queensland Department of Primary Industries, next to a CO60 irradiated accession of ‘Williams’ AAA that they developed. Although the selection resists race 4 in Queensland, Australia, it has succumbed here in a plot at the Taiwan Banana Research Institute (TBRI). The history of the banana improvement programs is filled with examples of "resistant" selections such as this that succumbed to Panama disease after they were disseminated to another region (photo courtesy of R. C. Ploetz). Fig. 9. Mike Smith (left) and Ken Pegg (right), of the Queensland Department of Primary Industries, next to a CO60 irradiated accession of ‘Williams’ AAA that they developed. Although the selection resists race 4 in Queensland, Australia, it has succumbed here in a plot at the Taiwan Banana Research Institute (TBRI). The history of the banana improvement programs is filled with examples of "resistant" selections such as this that succumbed to Panama disease after they were disseminated to another region (photo courtesy of R. C. Ploetz).
|
|
TR4 is distinguished from subtropical race 4 because it is genetically distinct and damages Cavendish in the tropics (30,31). Research is needed on the host ranges of the two pathotypes. Although they are similar, some clones, such as ‘Pisang Lilin’ AA, are affected only by TR4, and the reaction of other clones that are affected in the tropics, such as ‘Pisang Berangan’ AAA, is not known in the subtropics since they are not produced there. The tropical response of other clones that are affected in the subtropics is also not known (e.g., ‘Yangambi km 5’ AAA, ‘FHIA 03’ AABB, and ‘FHIA 23’ AAAA) (34).
Are There Additional Races of FOC?
Pathogenic diversity is not adequately defined with the current races of FOC (race 1 affects ‘Gros Michel,’ 2 ‘Bluggoe’ and 4 ‘Gros Michel,’ ‘Bluggoe’ and Cavendish) (28). For example, isolates from East Africa and Florida affect ‘Gros Michel’ and ‘Bluggoe’, but not Cavendish (Ploetz, unpublished; Stover, personal communication). New races might also be added as new susceptible cultivars are identified. ‘Hua Moa’ AAB (Fig. 10), a cultivar in the Maia Maoli-Popoulu subgroup, succumbed in recent field trials in Florida (33), and the differential susceptibility of ‘Pisang Lilin’ was noted above.
| |

Fig. 10. ‘Hua Moa’ AAB, a member of the Maia Maoli-Popoulu subgroup. These bananas are most important in the Pacific, and are very susceptible to Panama disease (photo courtesy of R. C. Ploetz). |
|
Research is also needed in other areas. The influence of environmental and edaphic conditions on the development of this disease is incompletely understood (43). In addition, the epidemiology of the TR4 outbreaks is confusing (30). The Cavendish epidemics have developed in plantations that were established with tissue-culture plantlets (supposedly pathogen-free) and in areas without a recent history of banana cultivation. Some of the relevant questions are: (i) How long can the pathogen survive in the absence of a banana host? (ii) What alternative, non-banana hosts are present in these production areas, and what is their distribution and impact? and (iii) If the tissue culture plantlets were pathogen-free, how was the pathogen moved to these production areas?
Resistance in Banana and Virulence in FOC
The above differentials are natural triploids whose parentage is unclear (see 5). Since their fertility is limited (Cavendish is sterile), traditional genetic analyses of their susceptibility and resistance have not been possible. Likewise, due to the absence of a sexual cycle in FOC, the genetic control of virulence is unexplored.
Work from the breeding programs indicates that several different genes are associated with resistance to Panama disease (summarized in 22). A single dominant gene was responsible for resistance (presumably to race 1) among tetraploid hybrids between ‘Gros Michel’ and various diploid parents, and a single dominant factor was associated with resistance to race 1 in progeny between three susceptible Musa taxa and ‘Pisang Lilin.’ In contrast, resistance to race 4 in a parent in the FHIA breeding program, ‘Pisang Juri Buaya’ AA, was thought to be polygenic.
Interestingly, recent work with a segregating population of a wild M. acuminata spp. malaccensis suggested that resistance to race 4 was due to a single recessive gene; molecular work indicated that it was homologous to the I2 gene that confers resistance in tomato to race 2 of F. oxysporum f. sp. lycopersici (25). Transformations are underway to determine whether this R-gene homolog will impart race 4 resistance to ‘Pome’ and a Cavendish cultivar, ‘Grand Nain.’
Phylogenetic Research on FOC
F. oxysporum is a complex of morphologically similar filamentous fungi. It is comprised of mainly saprophytic strains, but also contains plant pathogens that cause vascular wilts, rots and damping off of hundreds of host species (7,9,18). Agriculturally and economically, it is the most important species of Fusarium.
Plant-pathogenic members of F. oxysporum are classified in over 150 formae speciales, each of which has a unique host range of usually related taxa (2). Strains of F. oxysporum do not produce a teleomorph and generally cannot be distinguished morphologically (7,9,18).
Given the scant morphological features that are available, other characters have been important when examining variation and population structure in FOC (13,24,34). Significant progress became possible after the development of a relatively rapid method for determining VCGs (Fig. 11) (6,36).
| |
 |
|
| |
Fig. 11. Vegetative compatibility in FOC has been assessed with nitrate-nonutilizing, nit, mutants (6,34,36). (A) Wildtype isolates are cultured on a chlorate containing medium on which mutant sectors eventually form (lower righthand corner). (B) Mutant sectors that develop usually grow diffusely on a medium that contains nitrate as the sole source of N. These nit mutants are phenotyped for utilization of different nitrogen sources to determine the portion of the nitrate utilization pathway that has been affected. In (C) nit mutants have been plated on a basal medium that contains either ammonium (upper left), nitrate (upper right), hypoxanthine (lower left) or nitrite (lower right). Correll et al. (6) determined that the most reliable results in complementation tests were obtained when nit1 (nitrate reductase) and NitM (molybdenum cofactor, which is under the control of several genes) mutants were paired. The mutant at the top of each of these plates is a NitM mutant since it utilizes neither nitrate nor hypoxanthine. All that remain, except that on the lower left, utilize all but nitrate and are nit1 mutants. (D) Wild-type growth at the intersection of complementary mutants, as seen in the top of this picture, indicates vegetative compatibility (photos courtesy of R. C. Ploetz). |
|
For a fungus that has no known sexual stage, a surprising amount of diversity has been revealed by the vegetative compatibility work. To date, more than 20 VCGs of FOC have been reported (Table 1) (34). Some are distributed worldwide, whereas others have narrow distributions. Some of the VCGs have been recovered from a wide array of cultivars and genomes, some have come from specific banana genomes, and others have been found on a single cultivar. Subsequent phylogenetic work has improved our understanding of this variation.
Table 1. Vegetative compatibility among strains of Fusarium oxysporum f. sp. cubense.
| VCGx |
Genomic Group: Cultivar(s) |
Origin(s) |
0120-
01215 |
Musa sp.;
AA: SH-3142, SH-3362;
AAA: Gros Michel, Highgate, Pisang Ambon Putih, Pisang Ambon, Dwarf Cavendish, Williams, Mons Mari, Grand Nain, Lacatan;
AAB: Prata, Lady Finger, Pacovan, Hua Moa, Silk |
Australia, Brazil, Costa Rica, France (Guadeloupe, Guiana), Honduras, Indonesia (Java), Jamaica, Malaysia (Sarawak), Nigeria, Portugal (Madeira), South Africa, Spain (Canary Islands), Taiwan, USA (Florida) |
| 0121 |
AAA: Gros Michel, Cavendish |
Indonesia (Sumatra), Taiwan |
| 0122 |
AAA: Cavendish; ABB: Saba |
Philippines |
| 0123 |
AAA: Gros Michel, Grand Nain;
AAB: Silk, Latundan, Pisang Keling;
ABB: Pisang Awak, Kluai Namwa |
Malaysia (peninsular and Sarawak), Philippines, Taiwan, Thailand |
0124-
0125-
0128-
01220 |
AAA: Williams, Grand Nain;
AAAA: Jamaica 1242;
AAB: Lady Finger, Maçã, Manzano, Maqueño;
ABB: Pisang Awak, Ducasse, Kayinga, Zambia, Kluai Namwa, Bluggoe, Harare, Kholobowa, Dwarf Bluggoe, Mbufu, Burro Criolla, Pelipita, Ice Cream |
Thailand, Uganda, USA (Florida), Zaire |
| 0126 |
AA: Pisang Berlin;
AAA: Highgate;
AAB: Maqueño, Pisang Manurung |
Honduras, Indonesia (Irian Jaya, Sulawesi), Papua New Guinea, Philippines |
| 0129 |
AAA: Mons Mari;
AAB: Lady Finger |
Australia |
| 01210 |
AAA: Gros Michel;
AAB: Manzano |
Cayman Islands, Cuba, USA (Florida) |
| 01211 |
AA: SH-3142 |
Australia |
| 01212 |
AB: Ney Poovan;
AAB: Silk, Kisubi;
ABB: Pisang Awak, Bluggoe |
Tanzania |
01213-
01216 |
AA: Pisang Lilin, Pisang Mas;
AAA: Pisang Ambon,Valery, Williams, Grand Nain, Novaria, Red, Pisang Udang, Pisang Susu, Pisang Nangka, Pisang Barangan;
AAB: Pisang Raja Serah, Pisang Rastali, Pisang Rajah, Relong;
ABB: Pisang Awak, Pisang Awak Legor, Saba, Pisang Kepok, Pisang Caputu, Pisang Kosta;
Unknown: Pisang Batan |
Australia, Indonesia (Halmahera, Irian Jaya, Java, Sulawesi, Sumatra), Malaysia (peninsular), Taiwan |
| 01214 |
ABB: Harare, Mbufu |
Malawi |
| 01217 |
AAB: Pisang Rastali |
Malaysia |
| 01218 |
AAB: Pisang Rastali, Pisang Raja Serah;
ABB: Pisang Awak, Kluai Namwa, Pisang Kepok, Pisang Siam |
Indonesia (Java, Sumatra), Malaysia (peninsular), Thailand |
| 01219 |
AAA: Pisang Ambon, Pisang Ambon Putih;
Unknown: Pisang Raja Garing |
Indonesia (Java, Sumatra) |
| 01221 |
ABB: Kluai Namwa |
Thailand |
x VCGs were determined with nitrate auxotrophic nit mutants as described previously (6), and are summarized in reference 34. VCG complexes (e.g., 0120-01215) are those in which some isolates in different VCGs are capable of forming heterokaryons (26).
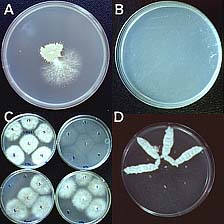
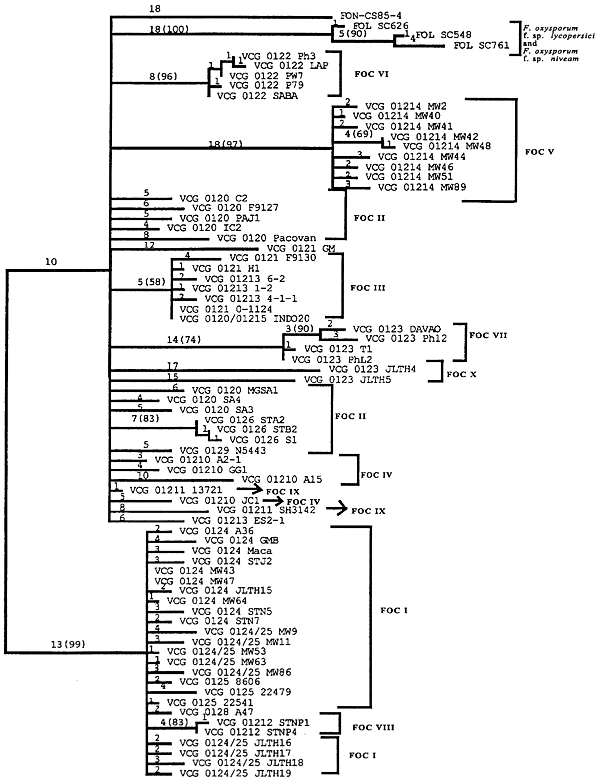
Koenig et al. (14) studied 165 strains of FOC in 13 VCGs with restriction fragment length polymorphisms (RFLPs) of genomic DNA and 19 anonymous, single-copy cDNA probes. Nine polymorphic RFLP loci and 72 unique genotypes were identified; only five of the genotypes accounted for half of the examined strains. Several clonal lineages were delineated with parsimony analysis (Fig. 12). Surprisingly, Lineage I (which corresponded to the VCG 0124-0125-0128 complex) and Lineage II (VCG 0120-01215) were genetically more similar to F. oxysporum f. sp. niveum, a pathogen of watermelon, than to each other, and were as closely related to each other as they were to F. oxysporum f. sp. lycopersici. Karyotype analyses have shown that chromosome number and genome size are significantly different in Lineages I and II (Fig. 13) (3,21).
|
|
|
|
Fig. 12. Midpoint-rooted 50% majority rule consensus tree representing 500 bootstrap replicates. One isolate represents each of the 72 restriction fragment length polymorphism haplotypes of Fusarium oxysporum f. sp. cubense, three isolates of F. oxysporum f. sp. lycopersici, and one isolate of F. oxysporum f. sp. niveum. Branch lengths are indicated on each branch, and bootstrap values are in parentheses. Tree length = 351, consistency index = 0.214, homoplasy index = 0.786, retention index = 0.705, and rescaled consistency index = 0.151. This figure appeared originally as Fig. 2 in reference 14. |
|
| |
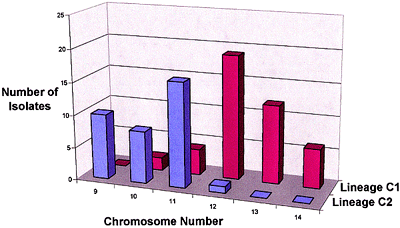
Fig. 13. Frequency distribution of chromosome numbers for isolates of Lineage C1 (corresponds to Lineage I of Koenig et al. (14) and the VCG 0124-0125-0128 complex) and C2 (Lineage II and VCG 0120-01215) of FOC (20). |
|
Lineage V (VCG 01214, which to date has been found only in the Misuku Hills in Malawi) was genetically distinct from all other FOC Lineages. Isolates in Lineage V affect enset, a musaceous plant that is endemic to Africa, as well as ‘Bluggoe’ (R. C. Ploetz, unpublished). Unlike other FOC Lineages that probably coevolved with banana in Southeast Asia, Lineage V may have evolved in Africa with this banana relative (30,32).
O’Donnell et al. (13,21) constructed genealogies for nuclear (beta-tubulin and translation elongation factor 1-alpha) and mitochondrial (mt small subunit ribosomal RNA) genes of 23 FOC isolates that represented unique genotypes identified by Koenig et al. (14), plus two or more individuals of F. oxysporum ff. spp. lycopersici, melonis and radicis-lycopersici. For the FOC isolates, there was complete agreement between the sequence phylogenies and those that were determined during the RFLP study. Parsimony analyses of the combined sequence data for the translation elongation factor 1-alpha and mt small subunit ribosomal RNA genes indicated that none of the examined ff. spp. were monophyletic (i.e., had common evolutionary origins) (21). This was the first time that ff. spp. of F. oxysporum were shown to be polyphyletic, a surprising conclusion given prior assumptions that members of a f. sp. should be closely related (13).
Subsequent phylogenetic studies have shown that the morphological species concept for F. oxysporum appears to be so broad that it is practically meaningless. Snyder and Hansen’s definition of F. oxysporum is polyphyletic, since it includes phylogenetically distinct taxa, such as F. redolens (Fig. 14) (13). Moreover, the forma specialis concept is phylogenetically misleading. Although monophyletic ff. spp. are known, multiple evolutionary histories are common (2,20). (Note that isolates of FOC in Fig. 14 fall into three independent clades.) This has important implications for understanding these important plant pathogens, and for the development and deployment of resistant genotypes of their hosts.
| |
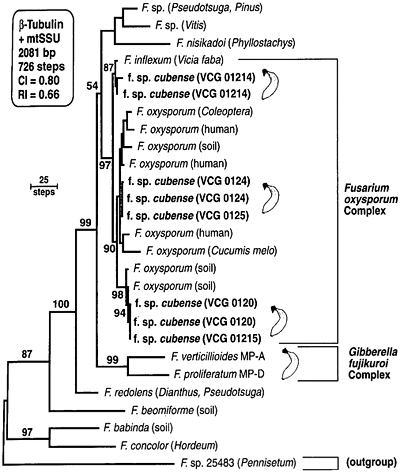
Fig. 14. A phylogram based on combined beta-tubulin and mtSSU rDNA sequence data for isolates of FOC. Numbers above nodes represent bootstrap intervals from 1000 replications. CI = consistency index, and RI = retention index. This figure appeared originally as Fig. 1 in reference 13. |
|
Since FOC has no known sexual cycle, it had been assumed to reproduce clonally. Koenig et al (14) provided genetic evidence to support this assumption. They showed that gametic disequilibrium among pairs of 34 of 36 RFLP alleles was significantly nonrandom. They also reported that strains with identical genotypes were found worldwide, and that the presence of independent genetic markers in these genotypes was strongly correlated. All of the above supported the notion that FOC reproduces clonally.
Taylor et al. (46) reanalyzed some of the RFLP data of Koenig et al. (14). They noted that in one of the largest clades identified by Koenig et al. (14) (it contained Lineages I and Lineages VIII in Fig. 12) there was little resolution and only one internal branch, which was consistent with recombination. Taylor et al. (46) used two different statistical tests, the index of association (IA) and the parsimony tree length permutation test (PTLPT), to determine whether alleles in this clade were recombining or were inherited clonally. By examining the 25 unique genotypes in the Lineage I + Lineage VIII clade they found no statistical support that its members reproduced in a strictly clonal fashion. Probabilities for the hypothesis that this population was significantly different from one that was recombining were 0.83 for the IA index and 0.85 for the PTLPT index (Figs. 15 and 16).
| |
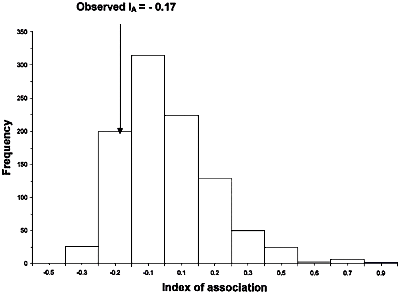
Fig. 15.
A title="" href="http://www.apsnet.org/publications/apsnetfeatures/Article%20Images/banana16.gif">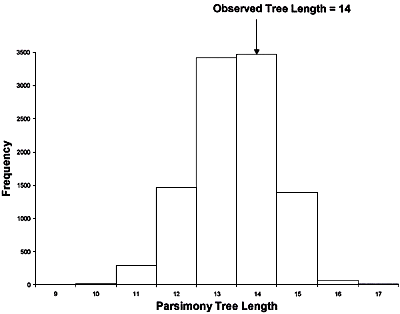
Fig. 16.
Reexamination of RFLP data from Koenig et al. (14). Taylor et al. (46) analyzed the 25 unique genotypes from FOC I and FOC VIII (see Fig. 12) with the Index of Association (IA) (Fig. 15) and parsimony tree length permutation test (PTLPT) (Fig. 16). Neither the observed IA (P = 0.83) nor the observed tree length (P = 0.85) are significantly different from what would be expected if the unique alleles in I and VIII were recombining. Figures are re-drawn from those that appeared in reference 46. |
|
These tests do not indicate what mechanisms of recombination are involved in Lineages I and VIII; parasexuality and sexual recombination may be operating. Although it is unclear whether such processes are ongoing or whether the results indicate historical recombination, it does appear that at least some members of FOC have diverse modes of reproduction. The extent to which this occurs in the Lineage I + VIII clade, whether it occurs in other members of FOC, and whether it occurs in other ff. spp. of F. oxysporum is not known but is relevant to our understanding of these pathogens and managing the important diseases that they cause.
Banana Improvement
Considering the long-term survival of FOC in infested soils, the absence of effective biological, chemical and physical control measures, and the susceptibility of many otherwise desirable cultivars, the development of new, resistant genotypes is of great importance (27,28).
The first banana-breeding program began in 1922 at the Imperial College of Tropical Agriculture (ICTA) in Trinidad (12,37). Its primary goal, and that of others that followed in Jamaica (Banana Board) and Honduras (Fundación Hondureña de Investigación Agrícola or FHIA), was to develop a Panama disease-resistant replacement for ‘Gros Michel.’ Another program in Brazil (Empresa Brasileira de Pesquisa Agropecuária or EMBRAPA) sought to develop Panama disease-resistant replacements for ‘Silk’ and ‘Pome.’
The breeding programs have had to overcome serious obstacles and in the early days (especially in Trinidad and Jamaica) had to start from scratch in several important areas (37). Nothing was known about the cytology and genetics of the genus Musa, its taxonomy was thoroughly confused, and collections of taxa that might be used in breeding did not exist. As the crop became better understood and collections were made in the Asian homeland of banana there were still major gaps in understanding which bananas were parents of the cultivated clones (5). Many of the desirable parents were either sterile (e.g., Cavendish) or if they did set seed were highly susceptibility (e.g., ‘Gros Michel’ and ‘Maqueño’). Moreover, the numbers and viability of seed that were set by the fertile parents were often very low and required the pollination of 1000s of bunches and embryo rescue to maximize the recovery of hybrids (Figs. 17 to 23). Currently, there are two important conventional breeding programs that focus on Panama disease.
| |

Fig. 17. |
|
| |

Fig. 18. |
|

Fig. 19. |
|
| |

Fig. 20. |
|

Fig. 21. |
|

Fig. 22. |
|

Fig. 23. |
The production of hybrids is similar in the various banana breeding programs. These pictures were taken at either the Fundación Hondureña de Investigación Agrícola (FHIA) in Honduras, or the International Institute of Tropical Agriculture (IITA) in Nigeria (the latter program has since relocated to Cameroon). Fig. 17. Female flowers are hand pollinated. Fig. 18. Mature, hybridized fruit are then ripened to facilitate seed extraction. Fig. 19. Fruit pulp is expressed with a mechanical press, Fig. 20, the black seed are removed and, Fig. 21, cleaned. To optimize the recovery of hybrids, embryo rescue is performed on the recovered seeds. Cleaned seeds are surface disinfested, their seed coats removed, and, Fig. 22, the embryos placed on a synthetic medium. Germinated embryos, Fig. 23, are ultimately transferred to the field for evaluation (photos courtesy of R. C. Ploetz).
The FHIA program, started by the United Fruit Company, has been in continuous operation since 1959. Located in La Lima, Honduras, its initial objective was the development of an export replacement, first for a Panama disease-resistant ‘Gros Michel’ and then for a black Sigatoka-resistant Cavendish. Emphasis is now also placed on other breeding targets such as the plantains and AAB dessert and ABB cooking bananas (12).
A major contribution of the FHIA program has been the development of synthetic diploid hybrids that are used as pollen parents, the SH lines (12). The SH parents are resistant to Panama disease, black Sigatoka and the burrowing nematode, Radopholus similis, and have played a key role in developing diverse tetraploid dessert and cooking hybrids. In general, the FHIA hybrids are high yielding and vigorous, but produce fruit with short post-harvest self-lives and organoleptic deficiencies. Work continues to improve the latter characteristics.
The EMBRAPA program began in an organized fashion in 1982 with the addition of the former, primary breeder in the Jamaican program, Kenneth Shepherd (12). It emphasized the development of Panama disease-resistant AAB dessert clones to replace ‘Maça’ (‘Silk’), ‘Prata’ (a Pome clone), and other susceptible cultivars. Resistance to black Sigatoka was subsequently added as a breeding target.
For the first time in the history of banana breeding, reasonable optimism exists that disease-resistant, widely acceptable clones will be available in the near future (Fig. 24). Unfortunately, many of the new hybrids are infected with Banana streak virus (BSV), a badnavirus that is integrated in the genome of diverse banana lines. BSV becomes episomal during meiosis and has appeared in hybrids among symptomless (virus free) parents. BSV has added a new, unexpected wrinkle to the development and dissemination of resistant genotypes (35).
| |

Fig. 24. Children in Africa, major beneficiaries of research on disease-resistant bananas (photo courtesy of R. C. Ploetz). |
|
Before concluding this article brief mention is made of two nonconventional approaches to banana improvement. A primary goal of the Taiwan Banana Research Institute (TBRI) in Pingtung, Taiwan has been the identification of race 4-resistant replacements for ‘Giant Cavendish’ (10). To achieve this goal, the GCTCVs have been selected in the field under high disease pressure and improved by recurrent selection. Some of the GCTCV selections produce acceptable yields of good fruit in Taiwan, but have not performed as well in subtropical areas, such as Australia and South Africa (Z. de Beer, personal communication; K. G. Pegg, personal communication). Some have been evaluated in the tropics, and are planted in Malaysia and Sumatra on a limited scale (10).
Several factors need to be considered when evaluating the performance of the GCTCV selections. Most important is the fact that banana is an annual crop in Taiwan, but grown as a perennial elsewhere (i.e., one to several ratoons are produced from the original plant crop). Moderate annual losses in Taiwan (5 to 15% per cycle for the best selections) increase rapidly to intolerable levels when they are compounded over several production cycles in perennial production systems. Thus, the acceptability of the GCTCV lines in perennial systems depends upon the number of cycles that are used.
Final mention is made of work to genetically transform banana for resistance to Panama disease (see 12 for a review). In general, progress with GM disease resistance in banana lags behind that that has been realized in other important food crops. This is not surprising. Genetic transformation and regeneration of banana are difficult and protocols for important genotypes were developed only recently. Furthermore, fungal diseases, which have been the most difficult to control in GM crops, are the most significant diseases of banana. Since the export companies conduct much of the work on banana, their proprietary results are generally unknown.
Whether export and local markets would accept fruit from disease-resistant transformants remains to be seen. Given the opposition to GM crops it is doubtful that GM bananas would be accepted in Europe in the near future. However, as consumers become aware of the benefits and safety of GM crops, this attitude will hopefully change. For example, a Cavendish clone that would resist TR4, as well as black Sigatoka and R. similis (problems for which the trades use massive amounts of pesticides), could increase subsistence production of these fruit and would immediately reduce the volumes of pesticides that are used in export production.
Conclusions
Due to its impact on the ‘Gros Michel’-based export trades, Panama disease was the most important disease of banana until 1960 (27,28,29,41). With the conversion to the Cavendish cultivars, the disease was no longer a problem in the tropical trades. It remained important only where Cavendish was affected in the subtropics and on other, less important cultivars.
The appearance of TR4 in Southeast Asia has changed this assessment (30,31). Once again the banana on which the export trades depend is threatened with destruction by Panama disease. Also affected are the important plantain cultivars that feed some 400 million people, as well as numerous cooking and dessert cultivars. In total, TR4 threatens more than 80% of the world’s banana production. Activities to monitor its spread and develop contingency plans for its arrival in other areas should be started as soon as possible.
The realization that many ff. spp. of F. oxysporum are comprised of two to several phylogenetically distinct taxa is recent but receiving increasing support (2,13,20). That some lineages of FOC are more closely related to other ff. spp. of F. oxysporum than to other members of FOC has direct relevance to understanding this pathogen and to the management of the important disease it causes. Associations between lineages and different banana cultivars represent, essentially, different pathosystems since the underlying genes for resistance and virulence may differ in each situation (13). The development of resistant genotypes, whether by conventional or nonconventional means, should recognize the variation in FOC that has been revealed by the phylogenetic work.
Literature Cited
1. APS. 2003. Plant pathologists unpeel rumors of banana extinction. Online. Press release, February 14. American Phytopathological Society, St. Paul, MN.
2. Baayen, R. P., O’Donnell, K., Bonants, P. J. M., Cigelnik, E., Kroon, L. P. N. M., Roebroeck, E. J. A., and Waalwijk, C. 2000. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 90:891-900.
3. Boehm, E. W. A., Ploetz, R. C., and Kistler, H. C. 1994. Statistical analysis of electrophoretic karyotype variation among vegetative compatibility groups of Fusarium oxysporum f. sp. cubense. Mol. Plant-Microbe Interact. 7:196-207.
4. Buddenhagen, I. W. 1990. Banana breeding and Fusarium wilt. Pages 107-113 in: Fusarium Wilt of Banana. R. C. Ploetz, ed. American Phytopathological Society, St. Paul, MN.
5. Carreel, F., Faur, J. S., Gonz lez de Len, D., Lagoda, P. J. L., Perrier, X., Bakry, F., Tezenas du Montcel, H., Lanaud, C., and Horry, J. P. 1994. Ivaluation de la diversité génétique chez les bananies diploVdes (Musa spp.). Genet. Sel. Evol. 26:125s-136s.
6. Correll, J. C., Klittich, C. J. R. and Leslie, J. F. 1987. Nitrate-nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology 77:1640-1646.
7. Domsch, K. H., Gams, W., and Anderson, T.-H. 1980. Compendium of Soil Fungi, Vol. 1. Academic Press, New York.
8. Gaidashova, S. V., Okech, S. H. O., Gold, C. S., and Nyagahungu, I. 2005. Why beer bananas? The case for Rwanda. InfoMusa 14:2-6.
9. Gerlach, W., and Nirenberg, H. 1982. The Genus Fusarium - a Pictorial Atlas. Paul Parey, Berlin, Germany.
10. Hwang, S.-C., and Ko, W.-H. 2004. Cavendish banana cultivars resistant to Fusarium wilt acquired through somaclonal variation in Taiwan. Plant Dis. 88:580-588.
11. International Network for the Improvement of Banana and Plantain. Online. INIBAP, a program of Consultative Group on Int. Agric. Res. (CGIAR) and Int. Plant Gen. Res. Inst. (IPGRI).
12. Jones, D. R., ed. 2000. Diseases of Banana, Abacá and Enset. CABI Publishing. Wallingford, UK.
13. Kistler, H. C. 2001. Evolution of host specificity in Fusarium oxysporum. Pages 70-82 in: Fusarium. Paul E. Nelson Memorial Symposium. B. A. Summerell, J. F. Leslie, D. Backhouse, W. L. Bryden, and L. W. Burgess, eds. American Phytopathological Society, St. Paul, MN.
14. Koenig, R., Ploetz, R. C., and Kistler, H. C. 1997. Fusarium oxysporum f. sp. cubense consists of a small number of divergent and globally distributed clonal lineages. Phytopathology 87:915-923.
15. Koeppel, D. 2005. Can this fruit be saved? Pop. Sci. 267:60-67, 104-105.
16. Kress, W. J. 1990. The phylogeny and classification of the Zingiberales. Ann. Missouri Botan. Garden 77:698-721.
17. Moore, N., Pegg, K. G., Langdon, P. W., Smith, M. K. and Whiley, A. W. 1993. Current research on Fusarium wilt of banana in Australia. Pages 270-284 in: Proceedings: International Symposium on Recent Developments in Banana Cultivation Technology, Taiwan Banana Research Institute, Chiuju, Pingtung, Taiwan, 14-18 December 1992. R. V. Valmayor, S. C. Hwang, R. C. Ploetz, S. W. Lee, and V. N. Roa, eds. INIBAP/ASPNET, Los Banos, Laguna, Philippines.
18. Nelson, P. E., Toussoun, T. A., and Marasas, W. O. 1983. Fusarium Species: An Illustrated Guide for Identification. Pennsylvania State University Press, University Park, PA.
19. New Agriculturist on-line. Points of view: The future of bananas. Online. WRENmedia, Fressingfield, Suffolk, UK.
20. O’Donnell, K., and Cigelnik, E. 1999. A DNA sequence-based phylogenetic structure for the Fusarium oxysporum species complex. Phytoparasitica 27:69.
21. O’Donnell, K. O., Kistler, H. C., Cigelnik, E., and Ploetz, R. C. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Nat. Acad. Sci. (USA) 95:2044-2049.
22. Ortiz, R. 1995. Musa genetics. Pages 84-109 in: Bananas and Plantains. S. Gowen, ed. Chapman & Hall, London, UK.
23. Pearce, F. 2003. Going bananas. New Sci. 177:26-29.
24. Pegg, K. G., Moore, N. Y. and Sorensen, S. 1994. Variability in populations of Fusarium oxysporum f. sp. cubense from the Asia/Pacific region. Pages 70-82 in: The Improvement and Testing of Musa: A Global Partnership, Proceedings of the First Global Conference of the International Musa Testing Program held at FHIA, Honduras, 27-30 April 1994. D. R. Jones, ed. INIBAP, Montpellier, France.
25. Pereza-Escheverria, S., Dale, J., Khanna, H., Smith, M., and Collet, C. 2004. Potential resistance gene against Fusarium wilt race 4. Pages 33 in: Abstract Guide. 1st International Congress on Musa. 6-9 July, 2004. Penang, Malaysia. INIBAP.
26. Ploetz, R. C., ed. 1990. Fusarium Wilt of Banana. American Phytopathological Society, St. Paul, MN.
27. Ploetz, R. C. 1992. Fusarium wilt of banana (Panama disease). Pages 270-282 in: Plant Diseases of International Importance, Vol. III. A. N. Mukhopadhyay, H. S. Chaube, J. Kumar, and U. S. Singh, eds. Prentice Hall, Englewood Cliffs, NJ.
28. Ploetz, R. C. 1994. Panama disease: Return of the first banana menace. Int. J. Pest Man. 40:326-336.
29. Ploetz, R. C. 2000. Panama disease: A classic and destructive disease of banana. Online. Plant Health Progress doi:10.1094/PHP-2000-1204-01-HM.
30. Ploetz R. 2004. Diseases and pests: A review of their importance and management. INFOMUSA 13:11-16.
31. Ploetz, R. C. Fusarium wilt of banana: A notorious disease that is caused by a phylogenetically diverse pathogen. Phytopathology (in review).
32. Ploetz, R. C., and Pegg, K. G. 1997. Fusarium wilt of banana and Wallace’s line: Was the disease originally restricted to his Indo-Malayan region? Australasian Plant Pathol. 26:239-249.
33. Ploetz, R. C., Vazquez, A., and Haynes, J. 1999. Responses of new banana accessions in South Florida to Panama disease. Crop Prot. 18:445-449.
34. Ploetz, R. C., and Pegg, K. G. 2000. Fusarium wilt. Pages 143-159 In: Diseases of Banana, Abacá and Enset. D. R. Jones, ed. CABI Publishing, Wallingford, UK.
35. Ploetz, R. C., Thomas, J. E. and Slabaugh, W. 2003. Diseases of Banana and Plantain. Pages 73-134 in: Diseases of Tropical Fruit Crops. R. C. Ploetz, ed. CABI Publishing, Wallingford, UK.
36. Puhalla, J. E. 1985. Classification of strains of Fusarium oxysporum on the basis of vegetative compatibility. Can. J. Bot. 63:179-183.
37. Shepherd, K. 1974. Banana research at I.C.T.A. Trop. Agric. (Trinidad) 51:482-490.
38. Simmonds, N. W., and Shepherd, K. 1955. Taxonomy and origins of cultivated bananas. J. Linneaen Soc. Bot. (London) 55:302-312.
39. Shivas, R. G., and Philemon, E. 1996. First record of Fusarium oxysporum f. sp. cubense on banana in Papua New Guinea. Australasian Plant Pathol. 25:260.
40. Shivas, R. G., Suyuko, S., Raga, N., and Hyde, K. D. 1996. Some disease-associated microorganisms on plants in Irian Jaya, Indonesia. Australasian Plant Pathol. 25:36-49.
41. Stover, R. H. 1962. Fusarial Wilt (Panama Disease) of Bananas and Other Musa species. CMI. Kew, Surrey, UK.
42. Stover, R. H. 1972. Banana, Plantain, and Abacá Diseases. CMI. Kew, Surrey, UK.
43. Stover, R. H., and Buddenhagen, I. W. 1986. Banana breeding: Polyploidy, disease resistance and productivity. Fruits 41:175-191.
44. Stover, R. H., and Simmonds, N. W. 1987. Bananas, 3rd ed. Longmans, London, UK.
45. Su, H. J., Hwang, S. C., and Ko, W. H. 1986. Fusarial wilt of Cavendish bananas in Taiwan. Plant Dis. 70:814-818.
46. Taylor, J. W., Jacobson, D. J., and Fisher, M. C. 1999. The evolution of asexual fungi: Reproduction, speciation and classification. Annu. Rev. Phytopathol. 37:197-246.
47. Thurston, H. D. 1997. Tropical Plant Diseases. 2nd ed. American Phytopathological Society, St. Paul, MN.
48. Waite, B. H. 1954. Vascular disease of abaca or Manila hemp in Central America. Plant Dis. Reptr. 38:575-578.
49. Waite, B. H. 1963. Wilt of Heliconia spp. caused by Fusarium oxysporum f. sp. cubense Race 3. Trop. Agric. (Trinidad) 40:299-305.
