Vidaver, A.K. and P.A. Lambrecht 2004. Bacteria as plant pathogens. The Plant Health Instructor. DOI: 10.1094/PHI-I-2004-0809-01
Anne K. Vidaver and Patricia A. Lambrecht
Department of Plant Pathology, University of Nebraska, Lincoln, NE
Introduction
Bacteria are single-celled microorganisms, generally ranging from 1-2 µm in size that cannot be seen with the unaided eye (Figure 1). Plant associated bacteria may be beneficial or detrimental. All plant surfaces have microbes on them (termed epiphytes), and some microbes live inside plants (termed endophytes). Some are residents and some are transient. Bacteria are among the microbes that successively colonize plants as they mature. Individual bacterial cells cannot be seen without the use of a microscope, however, large populations of bacteria become visible as aggregates in liquid, as biofilms on plants, as viscous suspensions plugging plant vessels, or colonies on petri dishes in the laboratory. For beneficial purposes or as pathogens, populations of 106 CFU (colony-forming units/milliliter) or higher are normally required for bacteria to function as biological control agents or cause infectious disease.
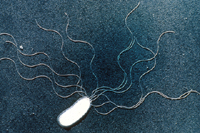
Figure 1 |
Plant pathogenic bacteria cause many serious diseases of plants throughout the world (Vidhyasekaran 2002; Figure 2), but fewer than fungi or viruses, and they cause relatively less damage and economic cost (Kennedy and Alcorn 1980). Most plants, both economic and wild, have innate immunity or resistance to many pathogens. However, many plants can harbor plant pathogens without symptom development (asymptomatic).

Figure 2 |
return to top
Frontiers: Current and Future Areas of Inquiry
The most exciting and current areas of research on plant-associated bacteria are the result of new intellectual discoveries, analyses and fields of study, new techniques and new instrumentation unavailable even a decade ago. For example, genomic sequencing, or the ordered reading of thousands of nucleotides constituting the deoxyribonucleic acid (DNA) of an organism, is now relatively common. Yet, of more than 166 sequenced and published bacterial genomes (National Center for Biotechnology Information/NCBI 2004) not including Archaea, at this time only eight are plant pathogens. Along with sequencing and its enormous data accumulation has arisen the field of bioinformatics to enable communication among scientists and analyses of the data, particularly comparative and evolutionary studies. Even supposedly simple steps like annotation of a gene, or its name or function, remains a challenge. The American Phytopathological Society has made a case of the need for additional bacteria to be sequenced (APS 2003). For maximal usefulness, such as unequivocal identification of an organism and determination of the number of its virulence genes and their locations, a fully annotated genome sequence is needed (Fraser et al. 2002). Analyzing the expression of DNA through intermediate steps with microarrays is a powerful emerging tool (Hinds et al. 2003). Similarly, methods for characterizing the entire protein complement of an organism (proteomics) are becoming available (Graves and Hayward 2002). These new and evolving techniques are enabling the study of virulence (the disease severity) and pathogenicity (the ability to cause disease), strain (descendants of a single isolation in culture) identification and typing (similarity or difference analysis relative to other strains), evolution and spread of bacteria, gene expression and gene regulation. These discoveries are occurring both with natural bacterial variants and mutants constructed in the laboratory. The expectation is to exploit these findings for improved disease management.
Mechanisms of pathogenicity of bacterial plant pathogens are becoming well known (Ahlemeyer and Eichenlaub 2001, Burger and Eichenlaub 2003). Virulence and pathogenicity genes may be harbored in different replicons (independent replicating units), such as spread throughout the chromosome(s) or in specialized areas termed genomic or pathogenicity islands (Arnold et al. 2003), in bacterial viruses integrated in the chromosome or in a 'carrier' state, and on one or more extra-chromosomal elements (plasmids). The functions of most genes, including those on extra-chromosomal elements, aren't known and it's estimated that each bacterium has about 40% of its genome devoted to unique genes.
Population development must normally occur for many bacteria to survive and infect plants. Infectious doses normally are in the millions of cells. In several cases, and perhaps all, the cells communicate chemically with one another (quorum sensing) and with other species. These chemical sensing molecules are under intensive study (Federle and Bassler 2003). In some cases, and perhaps most, microorganisms organize in dense growths to form biofilms that tightly adhere to surfaces, serving as protectants against the elements and enabling cells to produce a favorable environment for survival and spread.
Some structures used by bacteria to insert chemical compounds into plant cells are well studied, such as the so-called Type III secretion system (five types are currently known). The Type III system operates somewhat like a syringe and plunger to transport pathogen-produced proteins that effect disease or trigger defense (Pociano et al. 2003; Figure 3). These mechanisms have sometimes shown surprising and unexpected similarity to those found in animal and human pathogens (Cao et al. 2001). There are even a few strains of bacteria that cross kingdoms: they can infect both plants and humans. The genetic basis for such novelty is of immense interest and significance regarding the basis of infectious disease.
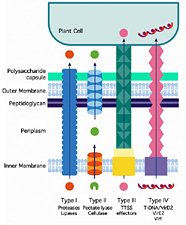
Figure 3 |
Commercial transgenic plants and those in development depend heavily on the use of a 'disarmed' pathogen, Agrobacterium tumefaciens, as a vector to insert a gene(s) of interest. Many challenges remain in transformation of certain plant varieties and species, as well as predictable and stable expression of transgenes (Gelvin 2003).
Challenges and opportunities for the future in plant microbiology abound (Vidaver 1999). The best is yet to come. For example, one of the current challenges is providing healthy plants for humans during long-term space travel and exploration (Ferl et al. 2002).
On the plant side, many avenues are being explored (Vidhyasekaran 2002). Understanding and manipulating resistance in host plants is extremely important. Host resistance may be due to one or several resistance genes (or R genes) to specific pathogens harboring virulence genes. If the virulence genes trigger a host defense response, they are termed avirulence (avr) genes. If the resistance is more general, a variety of preformed defense mechanisms, both structural and chemical, may be involved with induced chemicals as well (local or systemic acquired resistance) (Phuntumart 2003). Studies of pathogen interactions in model systems, particularly Arabidopsis thaliana, are enabling clearer understanding of susceptibility and resistance applicable to more complex plants (Heath 2002). Sequencing of major plant genomes is underway as well, with rice being completed. Multiple alleles and chromosomes, as well as complex traits are challenges in understanding and managing host resistance.
Compiling information from sequencing and functional analysis of both pathogens and major crop plants is expected to bring new insights useful for sustained disease management.
The forgoing depends on a basic knowledge of these bacteria, which follows.
return to top
History
Individual bacteria were first seen by humans about 325 years ago when they were magnified by the first microscope. It's only been a little over 100 years since a bacterium was first implicated as a causal agent in a plant disease. Bacteria were shown in 1878 to be associated with fireblight of apples and pears in Illinois and New York, USA (Burrill 1878). (fire blight disease lesson). The disease caused by Erwinia amylovora, now widespread throughout much of the temperate world, remains a limiting factor in growth of healthy apple and pear trees (Figure 4). In 1885, J.C. Arthur was able to isolate a bacterium from diseased plants, culture it, and then inoculate the same host to reproduce a naturally occurring disease. He recovered it subsequently from diseased tissue, fulfilling what is known as Koch's postulates (Arthur 1885). And it's only been about 120 years since the development of sterilized semi-solid media, first gelatin and then agar with various nutrients added, that enabled the isolation of purified cultures, a technique taken for granted today (Koch 1881).

Figure 4 |
Bacteria as plant pathogens can cause severe economically damaging diseases, ranging from spots, mosaic patterns or pustules on leaves (Figure 5) and fruits, or smelly tuber rots to plant death. Some cause hormone-based distortion of leaves and shoots called fasciation (Figure 6), or crown gall, a proliferation of plant cells producing a swelling at the intersection of stem and soil (Figure 7) and on roots.
Figure 5 |

Figure 6 |

Figure 7 |
return to top
Basic Biology
Bacteria associated with plants have several morphological shapes as can be seen with conventional microscopes at 400x to 1000x magnification. These shapes initially provided simple ways to differentiate them. There are bacilli (rods), cocci (spherical), pleomorphic rods (tendency toward irregular shapes) and spiral shapes. The majority of plant-associated bacteria are rods. However, modern science has shown by biochemical, genetic and molecular biological analyses that these bacteria are quite heterogeneous. Some are related to and grouped with animal and human pathogens.
By different types of microscopy (Basic Microscopy), principally fluorescent, confocal, phase-contrast and electron microscopy, one can see different parts of bacterial cells (Figure 8). Stains are often useful in the differentiation of structures.
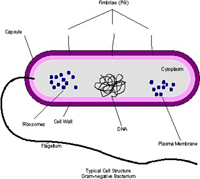
Figure 8 |
Chromosomes composed of DNA are coiled and there may be more than one per cell. Plasmids, or extra-chromosomal genomic entities may be present, and can code for essential virulence factors or conversely, biological control factors, which are chemicals effective against deleterious bacteria or fungi. Storage granules can be seen in some bacteria. Bacterial cells may or may not have appendages: flagella, usually at the poles of the cells (for movement) and fimbriae or pili, smaller thread-like appendages, usually at multiple locations. There is some evidence that flagellated cells produce larger lesions than non-flagellated mutants. The fimbriae are believed to be helpful in attachment, somewhat like a Velcro® fastener. Flagella and fimbriae, as well as different parts of the cell wall and cell membrane may contain receptor sites for bacterial viruses (bacteriophage) (Figure 9). However, bacteria without detectible appendages can be effective pathogens.
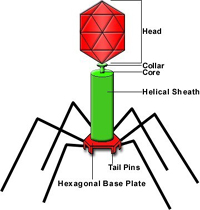
Figure 9 |
On laboratory media, plant pathogens usually grow more slowly than non-pathogenic bacteria isolated from plants, with optimal temperatures of 20-30°C (68-86°F). This makes isolation sometimes very challenging. A few grow at 37°C (99°F) (or higher), the temperature at which human pathogens, e.g. Burkholderia cepacia, (APSnet Feature: Burkholderia cepacia: Friend or Foe), are able to grow. Some can grow slowly at 10-12°C (50-54°F). Most are aerobic, some are facultative anaerobes (i.e. they can grow with or without oxygen), and a rare few are anaerobes. At high concentrations accompanying the growth of colonies (each colony is about 107 to 108 cells) on solid media, characteristic pigments may form within the colony or can be excreted into the growth medium, and may require special lighting (e.g. UV) for detection (Figure 10). Occasionally, bacterial pigments can be detected in seed (Figure 11). Medium requirements for growth may be simple or complex; some bacteria haven't been cultured. (Fastidious Vascular-Colonizing Bacteria). Some bacteria can produce characteristic volatile compounds, often with an unpleasant odor. Think of the smell of rotten potatoes, for example.
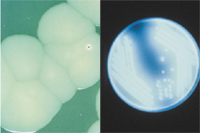
Figure 10 |

Figure 11 |
return to top
Reproduction
In general, bacteria reproduce by binary fission (one cell splitting into two), but the process is complex. If present, the extrachromosomal DNA elements or plasmids are usually reproduced in synchrony with the bacterial chromosome, but under some conditions can be lost naturally or by chemical manipulation ('cured'). Many plant pathogens harbor plasmids, e.g. strains of Pantoea (syn: Erwinia) stewartii subsp. stewartii (Stewart's wilt of corn disease lesson) may have up to 13 plasmids of unknown function (Coplin et al. 1981). Genetic variation in natural settings, e.g. fields, is probably underestimated due to lack of sampling and characterization. For example, at least seven pathogenic variants of the Goss's wilt and blight bacterium of corn, Clavibacter michiganensis subsp. nebraskensis (Smidt and Vidaver 1987), have been detected in a single field.
Horizontal transfer, the passage of DNA from one bacterial cell to another, may be accomplished in the laboratory and is assumed to account for much of the genetic variation among strains of a species and even among species in nature.
Bacteria may also harbor prophage, a stable inherited form of a bacterial virus or remnants of prophage integrated into their chromosomes. If a whole phage is integrated, some cells may lyse (break open) naturally or can be induced to lyse by chemical treatment, e.g. mitomycin C. In a very rare case, apparently both pathogenicity factors and a potent mammalian toxin are carried by a non-integrated bacterial virus in Rathayibacter toxicus (Ophel et al. 1993).
return to top
Systematics
Most of the plant pathogenic bacteria are either Gram-positive, classified within the Phylum Actinobacteria, or Gram-negative, in the Phylum Proteobacteria. Gram-positive and Gram-negative cells appear purple or red, respectively, with specific stains when viewed at 1000x magnification with a light microscope (Figure 12). The different colors largely reflect differences in stain retention by the respective cell walls of the bacteria during the staining process. Further differentiation is based on chemical or physiological characteristics, e.g. cell wall composition, enzyme production, substrate utilization, etc. Molecular characterization of 16S ribosomal RNA also may distinguish bacteria from one another. Ribosomes are coded by a highly conserved part of the bacterial chromosome and represent only a small part of the genome. But, the 'gold standard' for determining phylogenetic relationships is DNA:DNA homology by hybridization or genomic sequencing. Such analyses are sometimes at variance with ribosomal analyses.
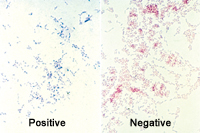
Figure 12 |
Interpretations of relationships vary with time, new techniques and more data. Thus, bacterial names can change from one era to another. It is sometimes challenging to read scientific literature and make legislative decisions without knowledge of all the synonyms for a particular bacterium. Updated nomenclature of bacterial plant pathogens can be found in Young et al. (1996) or accessed electronically in web-based bacterial nomenclature databases (Table 1), which are frequently updated. These listings include the historical and all published names for a particular bacterium.
In addition, due to the inability of humans to otherwise differentiate some host specific plant pathogenic bacteria, the concept of pathovars or pathogenic variants and races, differentiated by host range, can be found in the literature. This system is at variance with naming of pathogens of animals and humans, where host range differences may be recognized but are not part of an organism's name. Also, complicating the systematics of plant pathogenic bacteria is the presence of essential plasmids. Pathogenic Agrobacterium tumefaciens causes crown gall on a large number of hosts (crown gall disease lesson). Without its specific tumor-inducing plasmid (termed tumor-inducing or Ti plasmid) the strains are equivalent to nonpathogenic A. radiobacter.
return to top
Survival
Survival of plant pathogenic bacteria in nature occurs most commonly in plant debris left on the soil surface, in and on seeds, in soil, and in association with perennial hosts. But some bacteria can also survive in water and some do well on inanimate objects or on or inside insects. Clavibacter michiganensis subsp. sepedonicus, causative agent of potato ring rot, is notoriously known for surviving on machinery and packaging material. Knowledge of survival is usually essential to intervene in dissemination and for disease management.
return to top
Dissemination
Dissemination of plant pathogenic bacteria is easy, but fortunately does not always result in disease. Dissemination commonly occurs by windblown soil and sand particles that cause plant wounding, particularly during or after rains or storms (Figure 13). Wounding is essential for entry by many plant pathogens. Aerosols generated by diurnal temperature fluctuations enable dissemination, if temperature and humidity are aligned (Hirano and Upper 1989). Some plant diseases require certain temperature conditions e.g. Pseudomonas syringae (synonym: P. savastanoi) pv. phaseolicola causes disease below 22°C (72 °F) and Xanthomonas campestris (syn: X. axonopodis) pv. phaseoli, above 22°C on dry bean (Phaseolus vulgaris). Both diseases can occur simultaneously under growth conditions in which day and night temperature differentials enable disease progression in susceptible plants. Infested (surface contamination) or infected seed or any plant part can be sources of bacterial inoculum. Machinery, clothing, packing material and water can also disseminate pathogens, as can insects and birds. Continual monoculture in an area will usually enable increases in inoculum, making it easier for pathogens to be disseminated.
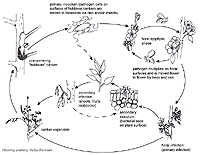
Figure 13 |
return to top
Host-Pathogen Interactions
Infection of plants by bacteria can occur in multiple ways. Infection is generally considered to be passive, i.e. accidental, although a few cases of plant chemoattractants have been reported. Bacteria can be sucked into a plant through natural plant openings such as stomata, hydathodes or lenticels. They can enter through abrasions or wounds on leaves, stems or roots or through placement by specific feeding insects. The nutrient conditions in plants may be such as to favor multiplication in different plant parts e.g. flowers or roots. Wind-driven rain carrying inoculum can be highly effective. Artificially, bacteria are most commonly introduced into plants by wounding, by pressure-driven aerosols mimicking wind-driven rains, vacuum infiltration, or by seed immersion into inoculum.
return to top
Symptomatology
Symptomatology of bacterial diseases is extremely varied, but usually characteristic for a particular pathogen. Symptoms can range from mosaics, resembling viral infections, to large plant abnormalities, such as galls or distorted plant parts. Hormone disruption can produce characteristic abnormal growths on roots, stems, and floral structures (phyllody) and sometimes abnormal flower colors (virescence). The most common symptoms are spots on leaves (Figure 14) or fruit (Figure 15), blights or deadening of tissue on leaves, stems or tree trunks, and rots (Figure16) of any part of the plant, usually roots or tubers. Wilts can also occur, due to plugging of vascular tissue (Figure 17). Symptoms may vary with photoperiod, plant variety, temperature and humidity, and infective dose. In some cases, symptoms may disappear or become inconsequential with further growth of the plant. For example, Holcus spot of corn caused by Pseudomonas syringae pv. syringae is arrested at the onset of hot dry weather.

Figure 14 |

Figure 15 |
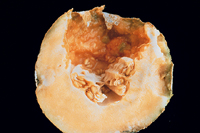
Figure 16 |

Figure 17 |
return to top
Diagnosis
Diagnosis of non-fastidious bacterial diseases depends on characteristic symptomatology, isolation of the presumed infectious agent, and physiological and/or molecular tests (Plant Disease Diagnosis). In heavily infected plants, bacterial populations in leaves or lesions may reach 108 or 109 CFU/gram of plant tissue, and actually visibly ooze from leaves or stems (Figure 18). A simple way to determine if a disease is caused by a bacterium is to cut a typical lesion or discolored area near its boundary with healthy tissue and suspend it in a droplet of water on a microscope slide. If a mass of moving small rods or 'dots' is seen at 400-1000x magnification flowing from the cut tissue under a microscope, you are observing bacterial streaming (Figure 19) which is an indicator of a bacterial disease. However, not all bacterial infections show streaming, or it may not be visualized without special microscope attachments. Serological tests, usually enzyme-linked, and physiological assays are available commercially for a few common and economically important bacteria. Molecular tests such as the polymerase chain reaction (PCR), based on specific genomic sequences, are becoming more readily available and used. Diagnostic tests are still evolving (Schaad et al. 2001), so that few are standardized and validated by multiple users, including governments.

Figure 18 |

Figure 19 |
Most plant pathogens are capable of inducing a hypersensitive reaction (HR) in plant species that are non-hosts or indicator plants (Klement et al. 1964). The HR is a plant defense mechanism elicited by the presence of a pathogen in non-host tissue. The tissue becomes sensitized to the pathogen, resulting in a rapid death of local plant cells (Figure 20), and entrapment of the pathogen. This, in effect, limits the spread of infection. One may use an HR test to determine if a colony isolated from infected plant tissue is a pathogen by introducing it, in a pure culture water suspension at 108 CFU/ml, into a non-host leaf panel. Tobacco (Nicotiana tabacum) is frequently used in HR tests because its large leaf panels are easily infiltrated, but Four O'clock (Mirabilis jalapa) may be used for some Gram-positive bacteria. Collapse of host tissue in the infiltration area within 48 hours indicates the bacterium is likely a pathogen for another host.

Figure 20 |
Confirmation that a pathogen causes disease symptoms requires a host and performance of a pathogenicity test. This strategy can be time-consuming (days, weeks, or months). A pure culture of bacteria recovered from diseased tissue is artificially inoculated into the same or related cultivar or another susceptible host species, in an effort to reproduce the same disease symptoms. The bacteria should then be reisolated and compared with the inoculant culture.
With some practice, most bacterial diseases can be easily diagnosed. However, the variation that can occur with different strains may require more sophisticated testing.
return to top
Epidemiology and Management
Bacterial diseases, in principle, can occur in any plant. Minimizing plant disease requires understanding the mechanisms of survival and spread. A competitive exclusion mechanism by beneficial bacteria can be effective in protection against disease. (Biological Control of Plant Pathogens). Notably, in crown gall of roses, Agrobacterium radiobacter strain K84 and its genetically engineered, transfer-minus derivative, strain K1026, provide excellent protection against A. tumefaciens (Ryder and Jones, 1991). Experimentally, and to a limited extent commercially, specific bacteriophages have been used as biological control agents and have merit based on having highly benign environmental effects.
In some cases, copper-based sprays are effective at minimizing inoculum build-up. But copper-resistant bacteria have also arisen. In a few cases, antibiotics used occasionally in human medicine have been used for combating plant diseases (Antibiotic Use for Plant Disease Management in the U.S.). Again, resistance to streptomycin has become quite common, although no resistance has yet been detected to tetracycline in plant pathogens (Vidaver 2002). No new antibiotics are likely to be available for use against plant pathogenic bacteria.
Producing clean seed is essential. Certification programs for some crops, e.g. potatoes, means initial plants are determined to be pathogen-free in tissue culture, from which growth is increased in greenhouses and then in strictly observed open field grow-out production before being released for general purchase. (blackleg of potato disease lesson) Burial of diseased debris is often useful along with crop rotation. Plant breeding for resistance (Breeding for Disease Resistance) is also often successful, if resistance is known to occur in some plant or can be projected to occur by transformation of certain genes, usually by vectors such as the Ti plasmid. These processes take several years and are specific to a particular plant cultivar. Eternal vigilance is necessary due to re-contamination potential, new pathogenic strains, and new susceptible hosts. On occasion, new pathogens are detected. Induced and acquired resistance can be obtained in some cases by the application of a chemical compound or other specific microorganisms to plants. This is a very active area of investigation.
return to top
Useful Plant Pathogens and Relatives
A few bacterial plant pathogens or their relatives have been widely used in agriculture and food production (Table 2). The thickening agent, xanthan gum, is an extra-cellular polysaccharide derived from the plant pathogen Xanthomonas campestris pv. campestris and is found in an enormous variety of products (Sutherland 1993). Transformation or genetic engineering of plants is best carried out by disarmed vectors (plasmids) of Agrobacterium tumefaciens. The elimination of a gene from a nonpathogenic Pseudomonas syringae that codes for ice formation at relatively high temperatures made history (Lindow 1987) in an ice-minus derivative that prevents frost damage when applied to plants. Other properties await discovery and exploitation.
Table 2.
Useful Plant Associated Bacteria |
| Taxon |
Function |
| Agrobacterium radiobacter K84 and K1026 |
Biological control |
| Agrobacterium sp. M4 |
Source of an experimental drug for cholesterol degradation |
| Agrobacterium radiobacter J14 |
Biodegradation of Atrazine, an agricultural herbicide |
| Agrobacterium tumefaciens |
Plasmid vector for plant transformation (genetic engineering) |
| Erwinia amylovora |
Source of harpin (Messenger), an elicitor of disease resistance in plants |
| Xanthomonas campestris pv. campestris |
Xanthan gum, a polysaccharide used in food production, agriculture, cosmetics and pharmaceuticals. |
| Several plant associated bacteria |
Restriction endonucleases, enzymes used for specific cutting of DNA in scientific research |
return to top
Acknowledgement
We thank J. Partridge for helpful discussion. This is a publication of the University of Nebraska.
return to top
References
Ahlemeyer, J. and Eichenlaub, R. 2001. Genetics of phytopathogenic bacteria. Prog. Bot.62: 98-113. (Gram-negative bacteria).
American Phytopathological Society. 2003. Microbial genomic sequencing. Perspectives of the American Phytopathological Society (revised 2003). 21 pp.
Arnold, D.L., Pitman, A., and Jackson, R.W. 2003. Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 4:407-420.
Arthur, J.C. 1885. Proof that the disease of trees known as pear blight is directly due to bacteria. N.Y. Agric. Exp. St. Bull. 2 n.s: 1-4.
Burger, A. and Eichenlaub, R. 2003. Genetics of phytopathogenic bacteria. Prog. Bot. 64:98-114. (Gram positive bacteria).
Burrill, Thomas Jonathan. 1878. Pear blight. Trans Ill. State Hort. Soc. 114-116.
Cao, H., Baldini, R.L. and Rahme, L.G. 2001. Common mechanisms for pathogens of plants and animals. Annu. Rev. Phytopathol. 39:259-284.
Coplin, D.L., Rowan, R.G., Chisholm, D.A. and Whitmoyer, R.E. 1981. Characterization of plasmids in Erwinia stewartii. Appl. Env. Microbiol. 42:599-604.
Federle, M.J. and Bassler, B.L. 2003. Interspecies communication in bacteria. J. Clin. Investig. 112:1291-1299.
Ferl, R., Wheeler, R., Levine, H.G. and Paul, A.L. 2002. Plants in space. Curr. Opin. Plant Biol. 5:258-263.
Fraser, C.M., Eisen, J.A., Nelson, K.E., Paulsen, I.T. and Salzberg, S.L. 2002. The value of complete microbial genome sequencing (You get what you pay for). J. Bacteriol. 184:6403-6405.
Gelvin, S.B. 2003. Agrobacterium-mediated plant transformation: the biology behind the "gene-jockeying" tool. Microbiol. Mol. Biol. Rev. 67:16-37.
Graves, P.R. and Haystead, T.A.J. 2002. Molecular biologist's guide to proteomics. Microbiol. Mol. Biol. Rev. 66:39-63.
Heath, M.C. 2002. Nonhost resistance in plants to microbial pathogens. Pages 47-57 in: Infectious Disease: Innate Immunity. R.A.B. Ezekowitz, J.A. Hoffmann, eds. Humana Press Inc., Totowa, NJ.
Hinds, J, Witney, A. and Vass, J.K. 2002. Microarray design for bacterial genomes. Methods Microbiol. 33: 67-82.
Hirano, Susan S. and Upper, Christen D. 1989. Diel variation in population size and ice nucleation activity of Pseudomonas syringae on snap bean leaflets. Appl. Env. Microbiol. 55:623-630.
Kennedy, B. W. and Alcorn, S.M. 1980. Estimates of U.S. crop losses to prokaryote plant pathogens. Plant Dis. 64:674-676.
Klement, Z., Farkas, G. L. and Lovrekovich, L. 1964. Hypersensitive reaction induced by phytopathogenic bacteria in the tobacco leaf. Phytopathology 54:474-477.
Koch, R. 1881. Zur untersuchung von pathogenen organismen. Mitth. a.d. Kaiserl. Gesundheitsampte 1: 1-48 In Milestones in Microbiology 1556 to 1940, translated and edited by Thomas D. Brock, ASM Press. 1998, p101.
Lindow, Steven E. 1987. Competitive exclusion of epiphytic bacteria by ice- Pseudomonas syringae mutants. Appl. Environ. Microbiol. 53:2520-2527.
National Center for Biotechnology Information, (NCBI), National Library of Medicine, National Institutes of Health, Bethesda, MD. Genome Specific Resources. 2004. http://www.ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html
Ophel, K.M., Bird, A.F. and Kerr, A. 1993. Association of bacteriophage particles with toxin production by Clavibacter toxicus, the causal agent of annual ryegrass toxicity. Phytopathology 83: 676-681.
Pierson, L.S. III and Ishimaru, C.A. 2000. Genomics of plant-associated bacteria: A glimpse of the future has become reality. APSnet Feature Article. Aug.1-Aug.31, 2000.
Phuntumart, V. 2003. Transgenic plants for disease resistance. Pages 180-215 in: Transgenic Plants: Current Innovations and Future Trends. C.N. Stewart, Jr., ed. Horizon Scientific Press, Wymondmam, UK.
Ponciano, G., Ishihara, H., Tsuyumu, S. and Leach, J.E. 2003. Bacterial effectors in plant disease and defense: keys to durable resistance? Plant Dis. 87:1271-1282.
Ryder, M.H. and Jones, D.A. 1991. Biological control of crown gall using Agrobacterium strains K84 and K1026. Aust. J. Plant Physiol. 18:571-579.
Schaad, N. W., Jones, J. B. and Chun, W. (ed.). 2001. Laboratory guide for identification of plant pathogenic bacteria. 3rd ed. American Phytopathological Society Press. St. Paul, MN.
Smidt, M.L. and Vidaver, A. K. 1987. Variation among strains of Clavibacter michiganense subsp. nebraskense isolated from a single popcorn field. Phytopathology 77: 388-392.
Sutherland, I. W. 1993. Xanthan. Pages 363-388 in: Xanthomonas. J. G. Swings and E.L. Civerolo. eds. Chapman & Hall, London, England.
Vidaver, A.K. 1999. Plant microbiology: Century of discovery, with golden years ahead. Am. Soc. Microbiol. News 65:358-363.
Vidaver, Anne K. 2002. Uses of antimicrobials in plant agriculture. Clin. Inf. Dis. 34 (Suppl 3): S107-S110.
Vidhyasekaran, P. 2002. Bacterial disease resistance in plants. Molecular biology and biotechnological applications. 452 pp. The Haworth Press, Binghamton, NY.
Young, J.M., Saddler, G.S., Takikawa, Y., DeBoer, S.H., Vauterin, L., Gardan, L., Gvozdyak, R. I. and Stead, D.E. 1996. Names of plant pathogenic bacteria 1864-1995. Rev. Plant Pathol. 75:721-763.
